Some protein sequences evolve more rapidly than others. But how can this be demonstrated? One approach is
Question:
Some protein sequences evolve more rapidly than others. But how can this be demonstrated? One approach is to compare several genes from the same two species, as shown for rat and human in the table. Two measures of rates of nucleotide substitution are indicated in the table.
Nonsynonymous changes refer to single-nucleotide changes in the DNA sequence that alter the encoded amino acid (for example, ATC → TTC, which gives isoleucine → phenylalanine). Synonymous changes refer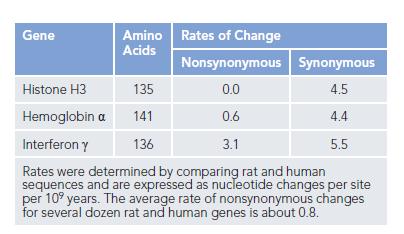
to those that do not alter the encoded amino acid (ATC→ ATT, which gives isoleucine → isoleucine, for example).
(As is apparent in the genetic code, Figure 7−27, there are many cases where several codons correspond to the same amino acid.)
A. Why are there such large differences between the synonymous and nonsynonymous rates of nucleotide substitution?
B. Considering that the rates of synonymous changes are about the same for all three genes, how is it possible for the histone H3 gene to resist so effectively those nucleotide changes that alter its amino acid sequence?
C. In principle, a protein might be highly conserved because its gene exists in a “privileged” site in the genome that is subject to very low mutation rates. What feature of the data in the table argues against this possibility for the histone H3 protein?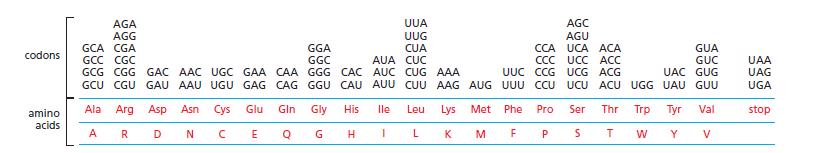
Step by Step Answer:

Essential Cell Biology
ISBN: 9780393680362
5th Edition
Authors: Bruce Alberts, Karen Hopkin, Alexander Johnson, David Morgan, Martin Raff, Keith Roberts, Peter Walter





