Answered step by step
Verified Expert Solution
Question
1 Approved Answer
4.6 Counting Amino Acids Protein sequences are made up of 20 different amino acids, each represented by a different letter of the alphabet. The valid
4.6 Counting Amino Acids
Protein sequences are made up of 20 different amino acids, each represented by a different letter of the alphabet. The valid letters are A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, and Y.
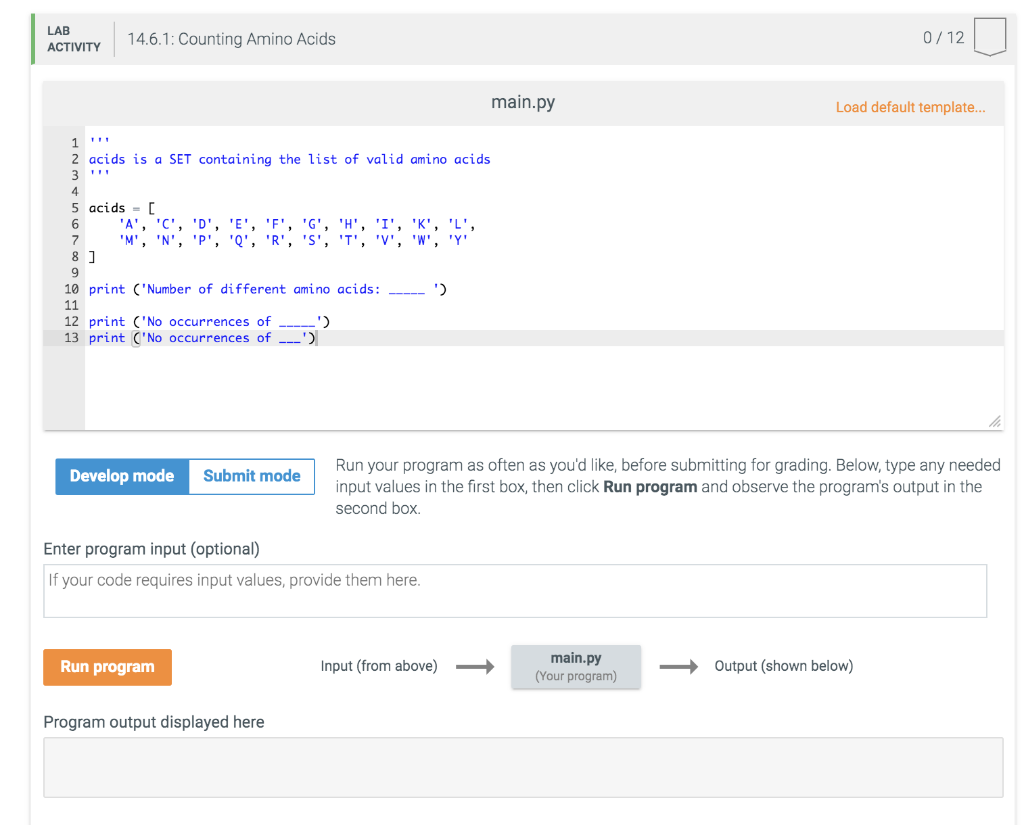
Write a program that reads in a protein sequence as a string and counts the number of each kind of amino acid in the sequence. Use a dictionary to accumulate the counts. Your program should not have a long series of "if/elif" statements!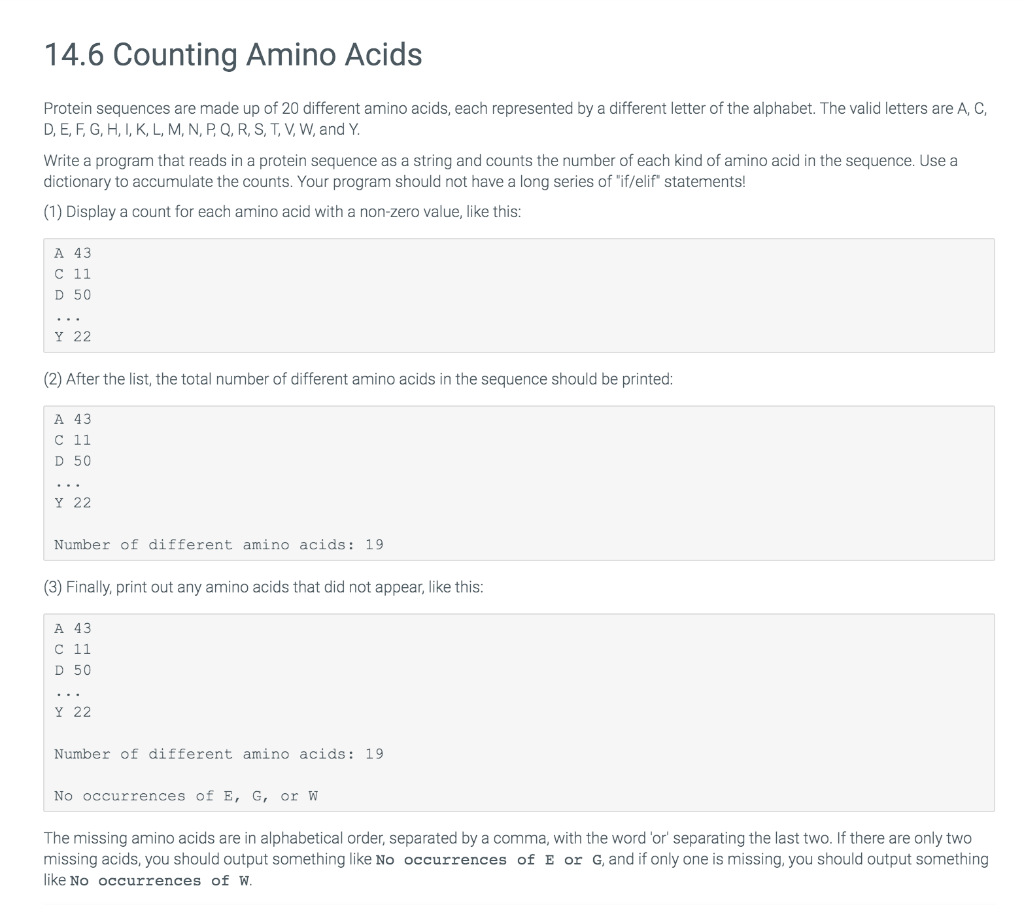
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


