Question
a) Given the DNA sequences S1, S2 and S3 and their alignments as below: $1 = TGCG S2 = AGCTG S3 = AGCG m
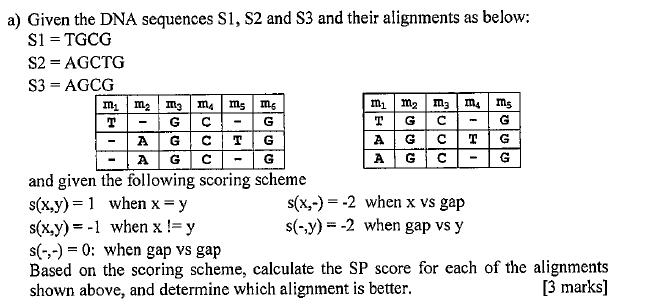
a) Given the DNA sequences S1, S2 and S3 and their alignments as below: $1 = TGCG S2 = AGCTG S3 = AGCG m m m T G C A G C A G MA m5 ms G G G and given the following scoring scheme s(x,y)=1 when x = y s(x,y) = -1 when x != y s(-,-) = 0: when gap vs gap Based on the scoring scheme, calculate the SP score for each of the alignments shown above, and determine which alignment is better. [3 marks] - m m m3 m4 ms T G G A G A G EUUU T EIHI s(x,-) = -2 when x vs gap s(-,y)=-2 when gap vs y T G G -
Step by Step Solution
There are 3 Steps involved in it
Step: 1
Step No 1 ANSWER To calculate the SP sumofpairs score for each alignment we need to assig...
Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get StartedRecommended Textbook for
Calculus
Authors: Ron Larson, Bruce H. Edwards
10th Edition
1285057090, 978-1285057095
Students also viewed these Programming questions
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
Question
Answered: 1 week ago
View Answer in SolutionInn App



