Question
Computational Phylogenetics Phylogenetics is the study of evolutionary history based on heritable traits such as DNA sequences. In computational phylogenetics (Links to an external site.)Links
Computational Phylogenetics
Phylogenetics is the study of evolutionary history based on heritable traits such as DNA sequences. In computational phylogenetics (Links to an external site.)Links to an external site., we use computer algorithms for this analysis. One of the key ideas in computational phylogenetics is to compare different sequences of DNA in order to calculate how different or similar two species are. Using this measure of genetic distance (Links to an external site.)Links to an external site., we can then determine how long ago a common ancestor would have lived. If two DNA sequences are very similar, a common ancestor could have lived recently; otherwise the common ancestor likely lived a longer time ago.
The following diagram illustrates how two species B and C have a very similar DNA and share a more recent ancestor while species A is very different from both B and C.
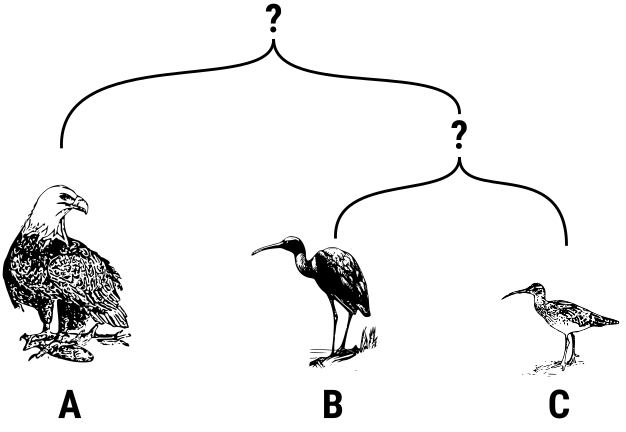
Your task is two determine which two out of three different DNA sequences are the most similar. The crucial insight is that DNA sequences are essentially just sequences composed of four different nucleobases. Therefore, we can represent DNA sequences in Python with strings that only use the four characters "A", "C", "G" and "T".
Step 1: Input and DNA Sequence Validation
First, the user needs to enter three different strings which correspond to three different DNA sequences A, B and C. You should ensure that the entered strings are actually valid DNA sequences. In particular, after the user enters the DNA sequences, you should validate that
all sequences include no characters other than "A", "C", "G" and "T", and
all sequences have the same length.
If the sequences entered by the user are not validated, you should print an error message and quit the program immediately with the statement "exit()".
Step 2: Calculating Genetic Distance
After the input sequences have been entered and validated, you should compute the genetic distance. There are different ways to determine the genetic distance between two DNA sequences. We will assume that the sequences always have the same length and use the Hamming Distance (Links to an external site.)Links to an external site. which compares two strings character-by-character and counts all the letters that are different. For example, the distance between "AAC" and "ACC" is 1 because the two strings differ in their second character. The distance between "AGG" and "GAA" is 3 because all three characters are different. Given the three strings from the user input, first determine the distance between A and B. You might want to use a for loop based on the length of A. As mentioned above, you can assume all strings have the same length. Then, you can copy and paste the same code and modify it to calculate the distance between A and C and between B and C.
Example: If A is "ACC", B is "CAT" and C is "GAT", then the distance between A and B is 3 (all characters are different), the distance between A and C is also 3 and the distance between B and C is 1 (because only the first character is different).
Step 3: Phylogenetic Tree Inference
The next step is to use the genetic distance matrix (Links to an external site.)Links to an external site. to reconstruct the evolutionary tree. In our case, we only have three different DNA sequences, so there can only be four different trees. We can determine the correct tree by finding the two out of these three species that are the most similar, so the two DNA sequences whose genetic distance is the smallest, the third sequence must therefore have an earlier common ancestor. As a special case, it is also possible that two or more distances are tied. In each of these cases, print an according message.
Example Interaction:
> DNA Sequence A: DNA Sequence B: DNA Sequence C: Species B and C have the most recent common ancestor.
A second example with longer DNA strings:
> DNA Sequence A: DNA Sequence B: DNA Sequence C: Species A and B have the most recent common ancestor.
(Here, lines starting with > indicate output from the program and lines starting with 2 2
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


