
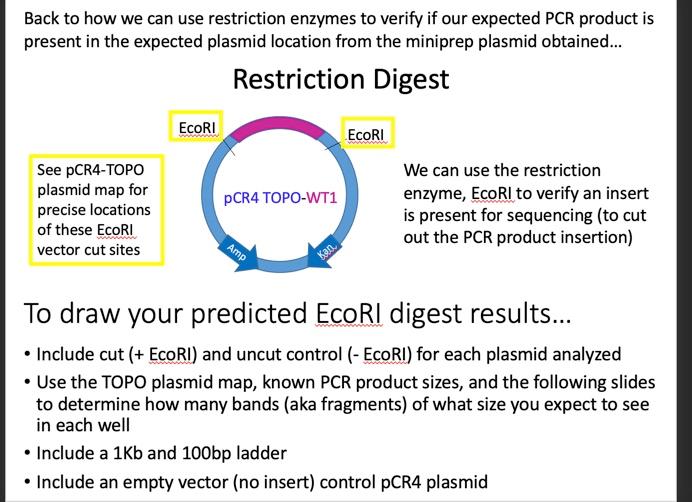
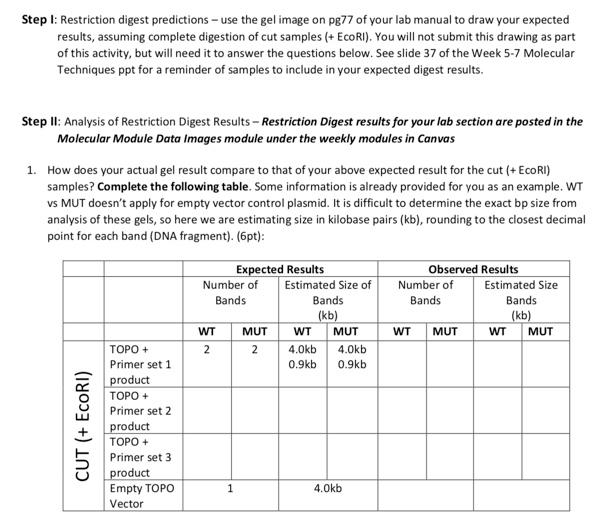
Copies of in-class activity sheets Data Analysis Activity 2 For this in-class activity, you will be working in your groups of 3-4 to analyze restriction digest results of your lab section's mini prep plasmids. Part 1: Restriction digest predictions 1. Draw your expected result on the gel image provided below for all conditions tested. Label wells for any samples analyzed in your section. 10kb Skb 3 b 2kb 1 db Ikb 0.5kb 0.5kb Part II: Analysis of Restriction Digest Results - answer for entire section's results 2. How does your actual gel result compare to that of your above predicted result? 3. Do you know from this analysis if all plasmids analyzed in your lab section contain the expected insert? Explain why or why not for each. 4. If any results are unexpected, can you suggest why? Please be sure to mind any updates changes to these activities to follow the version of the activity provided in-class 77 Back to how we can use restriction enzymes to verify if our expected PCR product is present in the expected plasmid location from the miniprep plasmid obtained... Restriction Digest EcoRI EcoRI pCR4 TOPO-WT1 See PCR4-TOPO plasmid map for precise locations of these EcoRI vector cut sites We can use the restriction enzyme, EcoRI to verify an insert is present for sequencing (to cut out the PCR product insertion) Amp Kan. To draw your predicted EcoRI digest results... Include cut (+ EcoRI) and uncut control (- EcoRI) for each plasmid analyzed Use the TOPO plasmid map, known PCR product sizes, and the following slides to determine how many bands (aka fragments) of what size you expect to see in each well Include a 1Kb and 100bp ladder Include an empty vector (no insert) control pCR4 plasmid Step 1: Restriction digest predictions - use the gel image on pg77 of your lab manual to draw your expected results, assuming complete digestion of cut samples (+ EcoRI). You will not submit this drawing as part of this activity, but will need it to answer the questions below. See slide 37 of the Week 5-7 Molecular Techniques ppt for a reminder of samples to include in your expected digest results, Step II: Analysis of Restriction Digest Results - Restriction Digest results for your la section are posted in the Molecular Module Data Images module under the weekly modules in Canvas 1. How does your actual gel result compare to that of your above expected result for the cut (+ EcoRI) samples? Complete the following table. Some information is already provided for you as an example. WT vs MUT doesn't apply for empty vector control plasmid. It is difficult to determine the exact bp size from analysis of these gels, so here we are estimating size in kilobase pairs (kb), rounding to the closest decimal point for each band (DNA fragment). (6pt): Expected Results Number of Estimated Size of Bands Bands (kb) WT MUT WT MUT 2 2 4.0kb 4.Okb 0.9kb 0.9kb Observed Results Number of Estimated Size Bands Bands (kb) WT MUT WT MUT CUT (+ EcoRI) TOPO + Primer set 1 product TOPO + Primer set 2 product TOPO + Primer set 3 product Empty TOPO Vector 1 4.Okb Copies of in-class activity sheets Data Analysis Activity 2 For this in-class activity, you will be working in your groups of 3-4 to analyze restriction digest results of your lab section's mini prep plasmids. Part 1: Restriction digest predictions 1. Draw your expected result on the gel image provided below for all conditions tested. Label wells for any samples analyzed in your section. 10kb Skb 3 b 2kb 1 db Ikb 0.5kb 0.5kb Part II: Analysis of Restriction Digest Results - answer for entire section's results 2. How does your actual gel result compare to that of your above predicted result? 3. Do you know from this analysis if all plasmids analyzed in your lab section contain the expected insert? Explain why or why not for each. 4. If any results are unexpected, can you suggest why? Please be sure to mind any updates changes to these activities to follow the version of the activity provided in-class 77 Back to how we can use restriction enzymes to verify if our expected PCR product is present in the expected plasmid location from the miniprep plasmid obtained... Restriction Digest EcoRI EcoRI pCR4 TOPO-WT1 See PCR4-TOPO plasmid map for precise locations of these EcoRI vector cut sites We can use the restriction enzyme, EcoRI to verify an insert is present for sequencing (to cut out the PCR product insertion) Amp Kan. To draw your predicted EcoRI digest results... Include cut (+ EcoRI) and uncut control (- EcoRI) for each plasmid analyzed Use the TOPO plasmid map, known PCR product sizes, and the following slides to determine how many bands (aka fragments) of what size you expect to see in each well Include a 1Kb and 100bp ladder Include an empty vector (no insert) control pCR4 plasmid Step 1: Restriction digest predictions - use the gel image on pg77 of your lab manual to draw your expected results, assuming complete digestion of cut samples (+ EcoRI). You will not submit this drawing as part of this activity, but will need it to answer the questions below. See slide 37 of the Week 5-7 Molecular Techniques ppt for a reminder of samples to include in your expected digest results, Step II: Analysis of Restriction Digest Results - Restriction Digest results for your la section are posted in the Molecular Module Data Images module under the weekly modules in Canvas 1. How does your actual gel result compare to that of your above expected result for the cut (+ EcoRI) samples? Complete the following table. Some information is already provided for you as an example. WT vs MUT doesn't apply for empty vector control plasmid. It is difficult to determine the exact bp size from analysis of these gels, so here we are estimating size in kilobase pairs (kb), rounding to the closest decimal point for each band (DNA fragment). (6pt): Expected Results Number of Estimated Size of Bands Bands (kb) WT MUT WT MUT 2 2 4.0kb 4.Okb 0.9kb 0.9kb Observed Results Number of Estimated Size Bands Bands (kb) WT MUT WT MUT CUT (+ EcoRI) TOPO + Primer set 1 product TOPO + Primer set 2 product TOPO + Primer set 3 product Empty TOPO Vector 1 4.Okb









