Answered step by step
Verified Expert Solution
Question
1 Approved Answer
DNA Analyser The program should implement the following high-level algorithm: Program: DNA Analyser Steps: 1 2 3 4 Display the program's name Prompt the
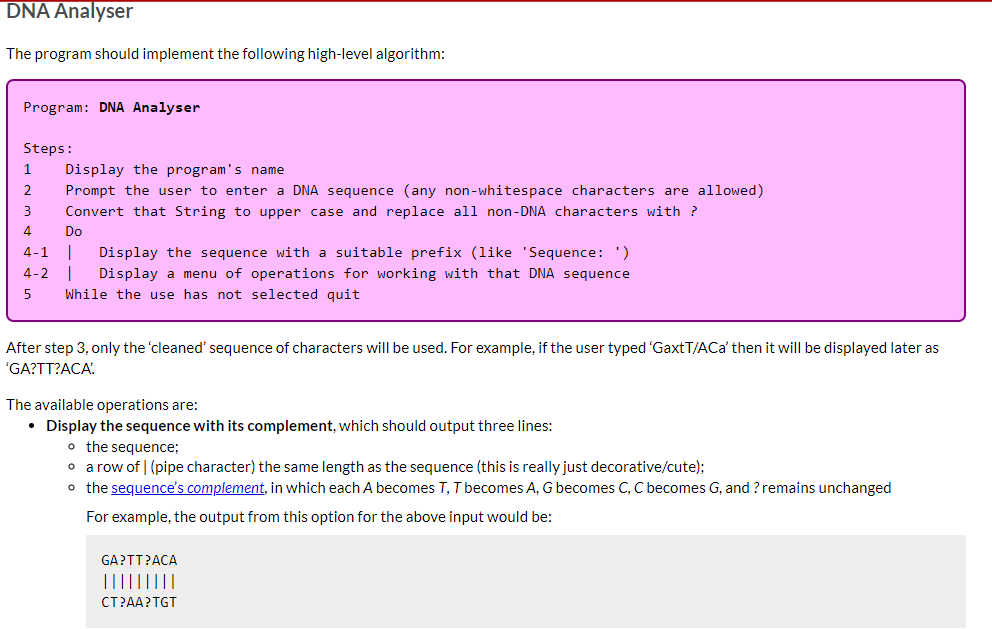

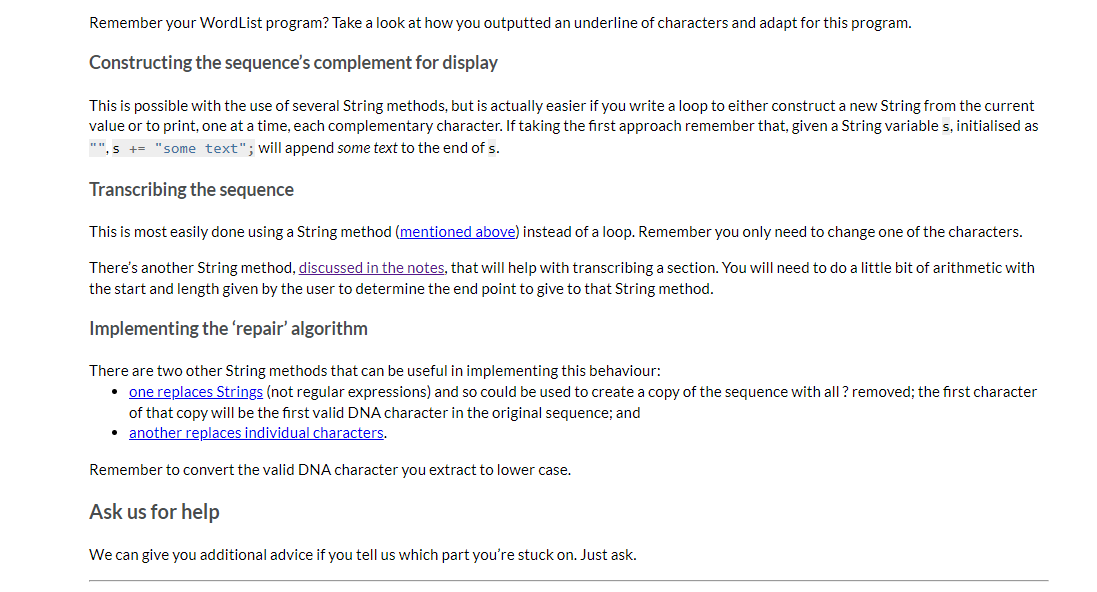
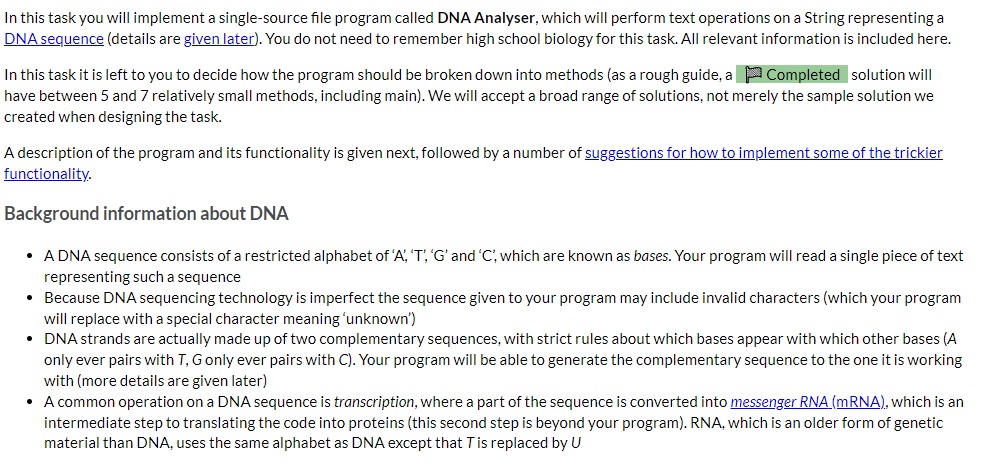
DNA Analyser The program should implement the following high-level algorithm: Program: DNA Analyser Steps: 1 2 3 4 Display the program's name Prompt the user to enter a DNA sequence (any non-whitespace characters are allowed) Convert that String to upper case and replace all non-DNA characters with ? Do 4-1 | Display the sequence with a suitable prefix (like 'Sequence: ') 4-2 | Display a menu of operations for working with that DNA sequence 5 While the use has not selected quit After step 3, only the 'cleaned' sequence of characters will be used. For example, if the user typed 'GaxtT/ACa' then it will be displayed later as 'GA?TT?ACA' The available operations are: Display the sequence with its complement, which should output three lines: o the sequence; o a row of | (pipe character) the same length as the sequence (this is really just decorative/cute); o the sequence's complement, in which each A becomes T, T becomes A, G becomes C, C becomes G, and ? remains unchanged For example, the output from this option for the above input would be: GA?TT?ACA ||||||||| CT?AA?TGT Transcribe the entire sequence, which should display the mRNA equivalent of the entire sequence (i.e., the sequence with all Ts converted to Us). Given the example above, the program would display 'GA?UU?ACA: Transcribe a section of the sequence. The user should be asked to give the start position (0-based) and length of the section to transcribe. Given the example above, if the user entered the start position 2 and length 5 the program would output '?UU?A? Show possible repair, which will display a version of the sequence in which all ?s have been replaced by a valid, lower case DNA character (the lower case indicates uncertainty over its value). It should apply the following rules (which are certainly not the way a recorded sequence would be repaired in real science): find the first valid DNA character in the sequence, convert that to lower case and then replace all? with that lower case character. There is advice below on how to achieve this using library methods and you can assume that there will be at least one valid DNA character in the user's input. Implementation advice Cleaning the input Strings have a replaceAll(String_regex, String_replacement) method that will replace all substrings that match the given regular expression (a pattern that can match a variety of String values) with the given replacement. As we don't cover regular expressions in the unit, here are some patterns that you will find useful: "[XYZ]" matches any single character from the set 'X', 'Y' or 'Z' "[^XYZ]" matches any single character that is not in the the set 'X', 'Y' or 'Z' You should not write a loop to clean the input text. Displaying the row of Is Remember your WordList program? Take a look at how you outputted an underline of characters and adapt for this program. Constructing the sequence's complement for display This is possible with the use of several String methods, but is actually easier if you write a loop to either construct a new String from the current value or to print, one at a time, each complementary character. If taking the first approach remember that, given a String variable s, initialised as Remember your WordList program? Take a look at how you outputted an underline of characters and adapt for this program. Constructing the sequence's complement for display This is possible with the use of several String methods, but is actually easier if you write a loop to either construct a new String from the current value or to print, one at a time, each complementary character. If taking the first approach remember that, given a String variable s, initialised as "", s += "some text"; will append some text to the end of s. Transcribing the sequence This is most easily done using a String method (mentioned above) instead of a loop. Remember you only need to change one of the characters. There's another String method, discussed in the notes, that will help with transcribing a section. You will need to do a little bit of arithmetic with the start and length given by the user to determine the end point to give to that String method. Implementing the 'repair' algorithm There are two other String methods that can be useful in implementing this behaviour: one replaces Strings (not regular expressions) and so could be used to create a copy of the sequence with all ? removed; the first character of that copy will be the first valid DNA character in the original sequence; and another replaces individual characters. Remember to convert the valid DNA character you extract to lower case. Ask us for help We can give you additional advice if you tell us which part you're stuck on. Just ask. In this task you will implement a single-source file program called DNA Analyser, which will perform text operations on a String representing a DNA sequence (details are given later). You do not need to remember high school biology for this task. All relevant information is included here. In this task it is left to you to decide how the program should be broken down into methods (as a rough guide, a Completed solution will have between 5 and 7 relatively small methods, including main). We will accept a broad range of solutions, not merely the sample solution we created when designing the task. A description of the program and its functionality is given next, followed by a number of suggestions for how to implement some of the trickier functionality. Background information about DNA A DNA sequence consists of a restricted alphabet of 'A', 'T', 'G' and 'C, which are known as bases. Your program will read a single piece of text representing such a sequence Because DNA sequencing technology is imperfect the sequence given to your program may include invalid characters (which your program will replace with a special character meaning "unknown') DNA strands are actually made up of two complementary sequences, with strict rules about which bases appear with which other bases (A only ever pairs with T, G only ever pairs with C). Your program will be able to generate the complementary sequence to the one it is working with (more details are given later) A common operation on a DNA sequence is transcription, where a part of the sequence is converted into messenger RNA (mRNA), which is an intermediate step to translating the code into proteins (this second step is beyond your program). RNA, which is an older form of genetic material than DNA, uses the same alphabet as DNA except that T is replaced by U
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


