Answered step by step
Verified Expert Solution
Question
1 Approved Answer
Hello i need help with the calculations and the drawing of the graphs. Basically the last picture where says laboratory report, the part of results.
Hello i need help with the calculations and the drawing of the graphs. Basically the last picture where says laboratory report, the part of results. 
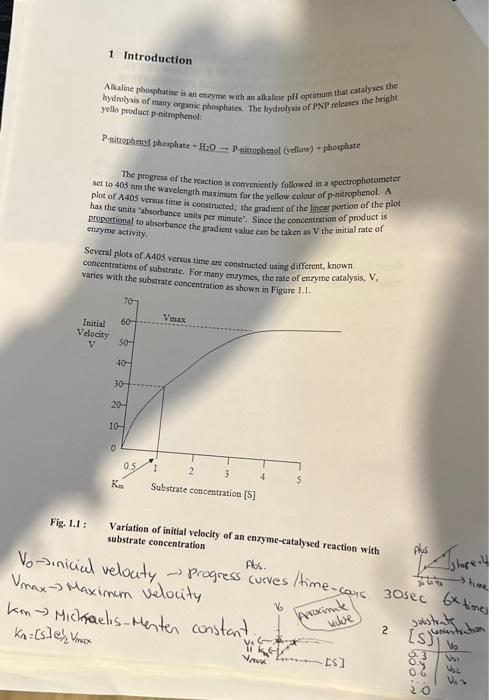
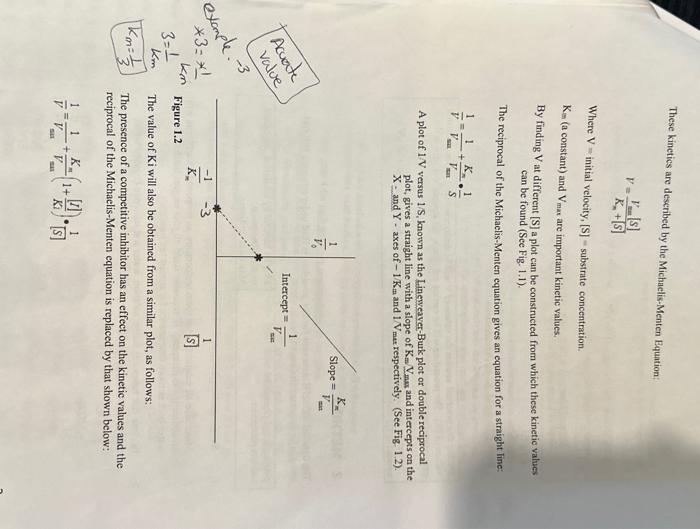



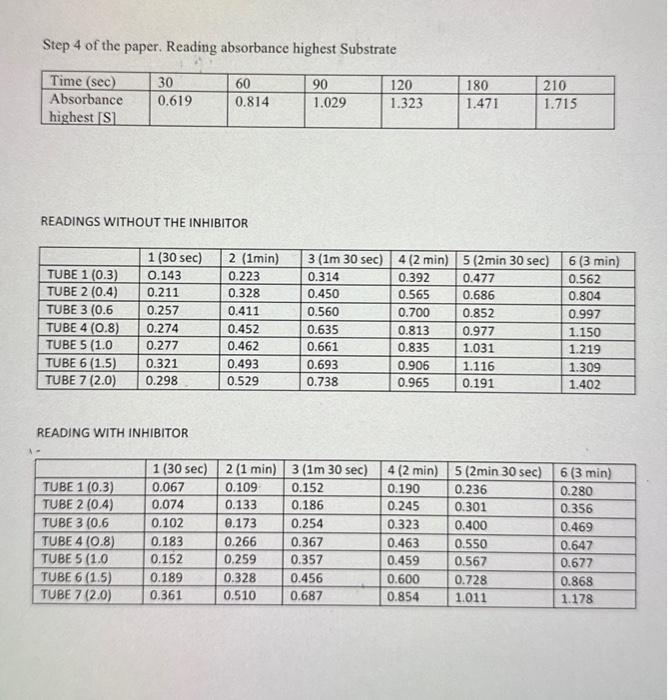
KINETICS OF A SINGLE SUBSTRATE ENZYME-CATALYSED REACTION; THE HYDROLYSIS OF P-NITROPHENYL PHOSPHATE BY MUCOSAL ALKALINE PHOSPHATASE Contents Introduction METHOD. LABORATORY REPORT FOR EXPERIMENT I 3.1 Results 3.2 Discussion .6 Note: Symbols and super and sub script may not show on mobile devices such as iPads. Alkaline phosphatise is an enzyme with an atkaline pH oftimutr that catalyses the bydmolysis of matio orsente phosphates, The bydrolysis of PNP releases the bright yello product penitrophenol: P.nitrombeny phosphate +H2O P.nitrophenol (yellon') + phosphate The progress of the reaction is conveniently followed in a spectrophotometer set to 405 nm the wavelength maximam for the yeliow colocr of p-nitrophenol. A plot of A405 versus time is constructed; the gmadient of the linear portion of the plod has the units 'absortuance units per minute'. Since the concentration of product is perpertiona1 to absorbance the per minute:. Since the concentration of product is enryme activity. Several plots of A405 versus time are constructed using different, known concentrations of substrate. For many enzymes, the rate of enzyme catalysis, V, varies with the substrate concentration as shown in Einuse i . Fig. 1.1 : Variation of initial velocity of an enzyme-catalysed reaction with substrate concentration Vo inicial velocity progress curves/time-cous 30 sec VmaxMaximamvelocity Macity These kinetics are deseribed by the Michaelis-Menten Equation V=Kn+[S]Vm+1[S] Where V= initial velocity, [S] - substrate concentration. K= (a constant) and Vmat are important kinetic values. By finding V at different [S] a plot can be constructed from which these kinetic values can be found (See Fig. 1.1). The reciprocal of the Michaelis-Menten equation gives an equation for a straight line: V1=Vsin1+VzunKnS1 A plot of 1V versus 1/S, known as the Lineweaver-Burk plot or double reciprocal plot, gives a straight line with a slope of KmVmax and intercepts on the X - and Y - axes of 1/Km and 1Vmat respectively. (See Fig 1,2). The value of Ki will also be obtained from a similar plot, as follows: The presence of a competitive inhibitor has an effect on the kinetic values and the reciprocal of the Michaelis-Menten equation is replaced by that shown below: V1=Vsin1+VminKm(1+Kl[I])[S]1 Where Ki is the dissociation constant for the enxyme - inhibitor complex and (I) is the concentration of the competitive inhibitor It can be seen that the slope of the plot is increased by a factor of 1+Kt[I] when a competitive inhibitor is present Slope of Iineweaver Burke plot "with inhibitor" Slope of L.ineweaver Burke plot "without inhibitot" =1+KI[I] Since [I] is 1mM, Ki can be calculated. During this practical session the values of V will be determined for a series of known values of [S]. The procedure will be carried out first in the absence and then in the presence of an inhibitor. The values of Vmax and Km will be obtained from a double reciprocal plot of 1/Vv versus 1/[S]. 2 METHOD The detailed COSHH statement for this procedure is filed under reference BFI/095 and can be inspected in Grenville room G209. 1. Set the spectrophotometer wavelength control to 405nm. 2. Make sure all solutions are at room temperature, as temperature variations will invalidate the experiment. If necessary stand all your reagent bottles in a water-bath at room temperature for at least ten minutes. 3. In clean test tubes mix buffer solution, water and substrate (5mM PNP in buffer) according to the following table, to give a range of selected values of [S]. Accurate pipetting is most important. 4. Starting with the highest substrate concentration transfer reaction solution to a cuvette, place the cuvette in the spectrophotometer and adjust for zero absorbance. Remove the cuvette, add 0.1ml of the enzyme solution and at the same time start a stopwatch and immediately mix the solution by inversion. Allow the cuvette to remain in the spectrophotometer until six readings have been taken. 5. If the reaction proceeds too rapidly for you to take readings (reaches an absorbance of 1.5 in less than one minute) your enzyme is too concentrated. Dilute the enzyme 1:1 with distilled water and try again with a fresh mixture. (Note that the spectrophotometer records absorbance values about 1.21.5 very badly). 6. When you are satisfied with the enzyme concentration, all subsequent measurements must be done with the same enzyme concentration. 7. Repeat step 4 for each substrate concentration in the table. 8. Draw graphs of A405 vs time (min) using the same set of axes for all values of [S]. Measure the gradient of the straight (linear) part of each graph. The gradient units are absorbance units per min. Take these values as your rates of activity. 9. Repeat the experiment, replacing the distilled water (0.1ml) with 0.1ml25 mM sodium phosphate to give a final concentration of 1.0mM sodium 3 LABORATORY REPORT FOR EXPERIMENT I Define clearly the aim(s) of the experiment. 3.1 Results - Clearly show [S]., V., 1/[S] and 1/V values in two tables - one showing values in the absence of inhibitor and one showing values in the presence of inhibitor. - Draw a Michaelis-Menten plot and a Lineweaver-Burk plot of your data. Use the same set of axes for each set of data i.e., with and without inhibitor. Use different symbols for the 2 sets of data e.g. O and . - For the Lineweaver-Burk plots draw a line of best fit through the points and from it determine the values of Vmax and Km with and without inhibitor. Use correct units; without units your values are meaningless, and credit cannot be given for your results. - Calculate the Ki of sodium phosphate. 3.2 Discussion Your discussion should include the following points: - Does your Lineweaver-Burk plot indicate that competition by sodium phosphate is competitive? - Comment on effect of the inhibitor on Km and Vmax values. - Explain the significance of Km and K i values. Step 4 of the paper. Reading absorbance highest Substrate READINGS WITHOUT THE INHIBITOR READING WITH INHIBITOR KINETICS OF A SINGLE SUBSTRATE ENZYME-CATALYSED REACTION; THE HYDROLYSIS OF P-NITROPHENYL PHOSPHATE BY MUCOSAL ALKALINE PHOSPHATASE Contents Introduction METHOD. LABORATORY REPORT FOR EXPERIMENT I 3.1 Results 3.2 Discussion .6 Note: Symbols and super and sub script may not show on mobile devices such as iPads. Alkaline phosphatise is an enzyme with an atkaline pH oftimutr that catalyses the bydmolysis of matio orsente phosphates, The bydrolysis of PNP releases the bright yello product penitrophenol: P.nitrombeny phosphate +H2O P.nitrophenol (yellon') + phosphate The progress of the reaction is conveniently followed in a spectrophotometer set to 405 nm the wavelength maximam for the yeliow colocr of p-nitrophenol. A plot of A405 versus time is constructed; the gmadient of the linear portion of the plod has the units 'absortuance units per minute'. Since the concentration of product is perpertiona1 to absorbance the per minute:. Since the concentration of product is enryme activity. Several plots of A405 versus time are constructed using different, known concentrations of substrate. For many enzymes, the rate of enzyme catalysis, V, varies with the substrate concentration as shown in Einuse i . Fig. 1.1 : Variation of initial velocity of an enzyme-catalysed reaction with substrate concentration Vo inicial velocity progress curves/time-cous 30 sec VmaxMaximamvelocity Macity These kinetics are deseribed by the Michaelis-Menten Equation V=Kn+[S]Vm+1[S] Where V= initial velocity, [S] - substrate concentration. K= (a constant) and Vmat are important kinetic values. By finding V at different [S] a plot can be constructed from which these kinetic values can be found (See Fig. 1.1). The reciprocal of the Michaelis-Menten equation gives an equation for a straight line: V1=Vsin1+VzunKnS1 A plot of 1V versus 1/S, known as the Lineweaver-Burk plot or double reciprocal plot, gives a straight line with a slope of KmVmax and intercepts on the X - and Y - axes of 1/Km and 1Vmat respectively. (See Fig 1,2). The value of Ki will also be obtained from a similar plot, as follows: The presence of a competitive inhibitor has an effect on the kinetic values and the reciprocal of the Michaelis-Menten equation is replaced by that shown below: V1=Vsin1+VminKm(1+Kl[I])[S]1 Where Ki is the dissociation constant for the enxyme - inhibitor complex and (I) is the concentration of the competitive inhibitor It can be seen that the slope of the plot is increased by a factor of 1+Kt[I] when a competitive inhibitor is present Slope of Iineweaver Burke plot "with inhibitor" Slope of L.ineweaver Burke plot "without inhibitot" =1+KI[I] Since [I] is 1mM, Ki can be calculated. During this practical session the values of V will be determined for a series of known values of [S]. The procedure will be carried out first in the absence and then in the presence of an inhibitor. The values of Vmax and Km will be obtained from a double reciprocal plot of 1/Vv versus 1/[S]. 2 METHOD The detailed COSHH statement for this procedure is filed under reference BFI/095 and can be inspected in Grenville room G209. 1. Set the spectrophotometer wavelength control to 405nm. 2. Make sure all solutions are at room temperature, as temperature variations will invalidate the experiment. If necessary stand all your reagent bottles in a water-bath at room temperature for at least ten minutes. 3. In clean test tubes mix buffer solution, water and substrate (5mM PNP in buffer) according to the following table, to give a range of selected values of [S]. Accurate pipetting is most important. 4. Starting with the highest substrate concentration transfer reaction solution to a cuvette, place the cuvette in the spectrophotometer and adjust for zero absorbance. Remove the cuvette, add 0.1ml of the enzyme solution and at the same time start a stopwatch and immediately mix the solution by inversion. Allow the cuvette to remain in the spectrophotometer until six readings have been taken. 5. If the reaction proceeds too rapidly for you to take readings (reaches an absorbance of 1.5 in less than one minute) your enzyme is too concentrated. Dilute the enzyme 1:1 with distilled water and try again with a fresh mixture. (Note that the spectrophotometer records absorbance values about 1.21.5 very badly). 6. When you are satisfied with the enzyme concentration, all subsequent measurements must be done with the same enzyme concentration. 7. Repeat step 4 for each substrate concentration in the table. 8. Draw graphs of A405 vs time (min) using the same set of axes for all values of [S]. Measure the gradient of the straight (linear) part of each graph. The gradient units are absorbance units per min. Take these values as your rates of activity. 9. Repeat the experiment, replacing the distilled water (0.1ml) with 0.1ml25 mM sodium phosphate to give a final concentration of 1.0mM sodium 3 LABORATORY REPORT FOR EXPERIMENT I Define clearly the aim(s) of the experiment. 3.1 Results - Clearly show [S]., V., 1/[S] and 1/V values in two tables - one showing values in the absence of inhibitor and one showing values in the presence of inhibitor. - Draw a Michaelis-Menten plot and a Lineweaver-Burk plot of your data. Use the same set of axes for each set of data i.e., with and without inhibitor. Use different symbols for the 2 sets of data e.g. O and . - For the Lineweaver-Burk plots draw a line of best fit through the points and from it determine the values of Vmax and Km with and without inhibitor. Use correct units; without units your values are meaningless, and credit cannot be given for your results. - Calculate the Ki of sodium phosphate. 3.2 Discussion Your discussion should include the following points: - Does your Lineweaver-Burk plot indicate that competition by sodium phosphate is competitive? - Comment on effect of the inhibitor on Km and Vmax values. - Explain the significance of Km and K i values. Step 4 of the paper. Reading absorbance highest Substrate READINGS WITHOUT THE INHIBITOR READING WITH INHIBITOR 






Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


