Answered step by step
Verified Expert Solution
Question
1 Approved Answer
in python..please follow rewuirements. program requirement. simple and clear write a function named clean_sequence() takes a string as its single parameter. It will use a
in python..please follow rewuirements.
program requirement. simple and clear
write a function named clean_sequence() takes a string as its single parameter. It will use a for loop to go through the string one character at a time; if the letter is alphabetic, convert it to upper case and add it on to the end of a result string. The function returns the result string. For example, clean_sequence('aaa, TCG, gaa') will return 'AAATCGGAA'
Use a while loop that goes through the cleaned sequence one triple at a time and processes it.

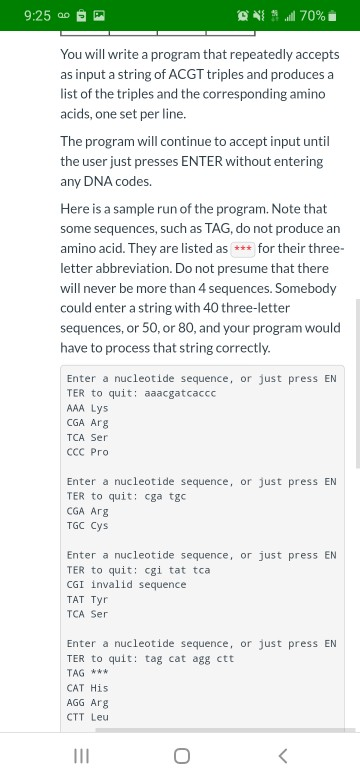

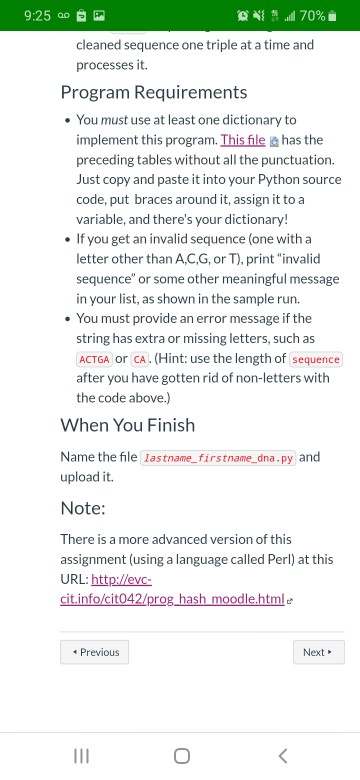
9:25 0 B ONUN 70% Dictionaries Submit Assignment Points 25 Due Tuesday by 11:59pm Submitting a file upload Read everything before doing anything! Background DNA is made up of sequences of nucleotides. There are four nucleotides: Adenine, Cytosine, Guanine, and Thymine, represented by the letters A, C, G, and T. Every set of three nucleotides produces one of twenty amino acids. For example, Cytosine, Thymine, and Guanine (CTG) produce Leucine. Note that there are 64 combinations of three nucleotides, and only twenty amino acids, so that means that a single amino acid can be produced in many different ways. The following tables, taken from the Biolnformatics tutoriale are Copyright 1999 by Boris Steipe, University of Munich. The table at the left shows the names of the amino acids and their abbreviations. The table at the right shows the abbreviations of the amino acids and the nucleotide sequences which produce them. | "T" "C" "G" "A" TTT:Phe TCT:Ser TGT:CyS TAT:Tyr TTC:Phe TCC:Ser TGC:Cys TAC:Tyr TTG:Leu TCG:Ser TGG:Trp TAG *** 9:25 20 ON 70% You will write a program that repeatedly accepts as input a string of ACGT triples and produces a list of the triples and the corresponding amino acids, one set per line. The program will continue to accept input until the user just presses ENTER without entering any DNA codes. Here is a sample run of the program. Note that some sequences, such as TAG, do not produce an amino acid. They are listed as *** for their three- letter abbreviation. Do not presume that there will never be more than 4 sequences. Somebody could enter a string with 40 three-letter sequences, or 50, or 80, and your program would have to process that string correctly. Enter a nucleotide sequence, or just press EN TER to quit: aaacgatcaccc AAA Lys CGA Arg TCA Ser ccc Pro Enter a nucleotide sequence, or just press EN TER to quit: cga tgc CGA Arg TGC Cys Enter a nucleotide sequence, or just press EN TER to quit: cgi tat tca CGI invalid sequence T Tyr CA Ser Enter a nucleotide sequence, or just press EN TER to quit: tag cat agg ctt TAG WWW CAT His AGG Arg CTT Leu 9:25 0 B ON 70% i Enter a nucleotide sequence, or just press EN TER to quit: tag cat agg ctt TAG *** CAT His AGG Arg CTT Leu Enter a nucleotide sequence, or just press EN TER to quit: tca cag gtt cg Error: You must give complete triples. Enter a nucleotide sequence, or just press EN TER to quit: Breaking a String into Triples This takes a bit of doing. I suggest you do this: Write a function named clean_sequence takes a string as its single parameter. It will use a for loop to go through the string one character at a time; if the letter is alphabetic, convert it to upper case and add it on to the end of a result string. The function returns the result string. For example, clean_sequence('aaa, TCG, gaa') will return 'AAATCGGAA' Use a while loop that goes through the cleaned sequence one triple at a time and processes it. Program Requirements . You must use at least one dictionary to implement this program. This file has the preceding tables without all the punctuation. Just copy and paste it into your Python source code, put braces around it, assign it to a variable, and there's your dictionary! . If you get an invalid sequence (one with a INT 9:25 PM ONS 70% cleaned sequence one triple at a time and processes it. Program Requirements . You must use at least one dictionary to implement this program. This file has the preceding tables without all the punctuation. Just copy and paste it into your Python source code, put braces around it, assign it to a variable, and there's your dictionary! If you get an invalid sequence (one with a letter other than A,C,G, or T), print "invalid sequence" or some other meaningful message in your list, as shown in the sample run. . You must provide an error message if the string has extra or missing letters, such as ACTGA OG CA. (Hint: use the length of sequence after you have gotten rid of non-letters with the code above.) When You Finish Name the file lastname_firstname_dna.py and upload it. Note: There is a more advanced version of this assignment (using a language called Perl) at this URL: http://evc- cit.info/cit042/prog hash moodle.html Previous Next
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


