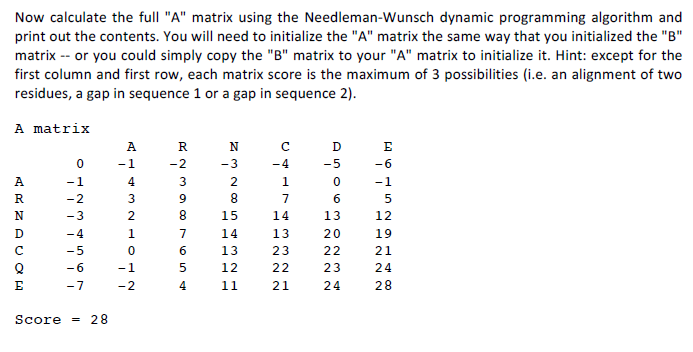
Question
MATLAB BIOINFORMATICS HOMEWORK HELP NEED HELP WITH PART 2 MAINLY. start code with this: % A trial sequence with all amino acids: % ARNDCQEGHILKMFPSTWYV %
MATLAB BIOINFORMATICS HOMEWORK HELP
NEED HELP WITH PART 2 MAINLY.
start code with this:
% A trial sequence with all amino acids:
% ARNDCQEGHILKMFPSTWYV
% The trial sequences in the HW description:
% ARNDCQE
% ARNCDE
% Ask the user for protein sequence 1.
prompt = 'Enter the first amino acid sequence of length 1 to 25: ';
seq1 = input(prompt, 's');
len1 = length(seq1);
fprintf(1, 'Seq 1 %s has %d residues ', seq1, len1);
% Ask the user for protein sequence 2.
prompt = 'Enter the second amino acid sequence of length 1 to 25: ';
seq2 = input(prompt, 's');
len2 = length(seq2);
fprintf(1, 'Seq 2 %s has %d residues ', seq2, len2);
% Initialize a 25x25 matrix of zeros.
A = zeros(25,25);
% The supplied initMatrix must be modified!
A = initMatrix(A, seq1, seq2);
% Print the matrix A.
printMatrix(A, seq1, seq2);
end
% Print out a matrix
function mat = initMatrix(mat, seq1, seq2)
numRows = length(seq1) + 1;
numCols = length(seq2) + 1;
mat(1,1) = 0; % Init the value at (1,1) to 0.
% Initialize the first column.
for i=2:numRows
% Do something here!
end
% Initialize the first row.
for c=2:numCols
% Do something here!
end
% Init the rest of the matrix.
for r=2:numRows
for c=2:numCols
% Do something here!
end
end
end
% Print out a matrix
function printMatrix(mat, seq1, seq2)
len1 = length(seq1);
len2 = length(seq2);
% Print the first row of sequence letters
fprintf(1,'\t\t');
for r=1:len2
fprintf(1,'%2c\t', seq2(r));
end
fprintf(1,' ');
% Print the first row of scores.
fprintf(1,'\t');
for c=1:len2+1
fprintf('%2d\t', mat(1,c));
end
fprintf(1,' ');
% Print the rest of the scores.
for r=2:len1+1
% Print the first letter of vertical sequence
fprintf(1, '%c\t', seq1(r-1));
for c=1:len2+1
fprintf(1,'%2d\t',mat(r,c));
end
fprintf(1,' ');
end
end
function value = scorePair(char1, char2)
% Order: ARNDCQEGHILKMFPSTWYV
i1 = aaToInt(char1);
i2 = aaToInt(char2);
A = [
4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -1 -1 -2 -1 1 0 -3 -2 0;
- 1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3;
- 2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3;
- 2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3;
0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1;
- 1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 -1 -2;
- 1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2;
0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3;
- 2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -1 -2 -1 -2 -2 2 -3;
- 1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3;
- 1 -2 -3 -4 -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 -2 -1 1;
- 1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2;
- 1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 -1 -1 1;
- 2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1;
- 1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2;
1. -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2;
0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5 -2 -2 0;
- 3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11 2 -3;
- 2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1;
0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4];
value = A(i1, i2);
end
function value = aaToInt(aa)
switch aa
case 'A',
value = 1;
case 'R',
value = 2;
case 'N',
value = 3;
case 'D',
value = 4;
case 'C',
value = 5;
case 'Q',
value = 6;
case 'E',
value = 7;
case 'G',
value = 8;
case 'H',
value = 9;
case 'I',
value = 10;
case 'L',
value = 11;
case 'K',
value = 12;
case 'M',
value = 13;
case 'F',
value = 14;
case 'P',
value = 15;
case 'S',
value = 16;
case 'T',
value = 17;
case 'W',
value = 18;
case 'Y',
value = 19;
case 'V',
value = 20;
otherwise,
value = 1;
end
end

Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


