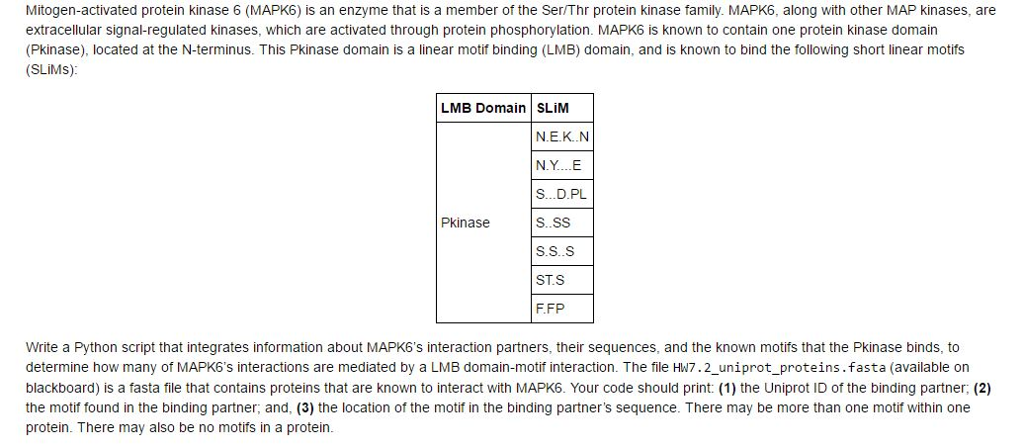
Mitogen-activated protein kinase 6 (MAPK6) is an enzyme that is a member of the Ser/Thr protein kinase family. MAPK6, along with other MAP kinases, are extracellular signal-regulated kinases, which are activated through protein phosphorylation. MAPK6 is known to contain one protein kinase domain (Pkinase), located at the N-terminus. This Pkinase domain is a linear motif binding (LMB) domain, and is known to bind the following short linear motifs (SLiMs); Write a Python script that integrates information about MAPK6's interaction partners, their sequences, and the known motifs that the Pkinase binds, to determine how many of MAPK6's interactions are mediated by a LMP domain-motif interaction. The file HW 7.2_uniprot_proteins.fasta (available on blackboard) is a fasta file that contains proteins; that are known to interact with MAPK6. Your code should print: (1) the Uniprot ID of the binding partner; (2) the motif found in the binding partner, and, (3) the location of the motif in the binding partner's sequence. There may be more than one motif within one protein. There may also be no motifs in a protein. Mitogen-activated protein kinase 6 (MAPK6) is an enzyme that is a member of the Ser/Thr protein kinase family. MAPK6, along with other MAP kinases, are extracellular signal-regulated kinases, which are activated through protein phosphorylation. MAPK6 is known to contain one protein kinase domain (Pkinase), located at the N-terminus. This Pkinase domain is a linear motif binding (LMB) domain, and is known to bind the following short linear motifs (SLiMs); Write a Python script that integrates information about MAPK6's interaction partners, their sequences, and the known motifs that the Pkinase binds, to determine how many of MAPK6's interactions are mediated by a LMP domain-motif interaction. The file HW 7.2_uniprot_proteins.fasta (available on blackboard) is a fasta file that contains proteins; that are known to interact with MAPK6. Your code should print: (1) the Uniprot ID of the binding partner; (2) the motif found in the binding partner, and, (3) the location of the motif in the binding partner's sequence. There may be more than one motif within one protein. There may also be no motifs in a protein







