Question
Please help. In my program, I am trying to look for the start codons(start codon = ATG) and stop codons(TAG, TAA, and TGA). My code
Please help. In my program, I am trying to look for the start codons(start codon = ATG) and stop codons(TAG, TAA, and TGA).


My code so far:
Part of the fna file:
>NC_003988.1 Simian enterovirus A, complete genome GAGTGTTCCCACCCAACAGGCCCACTGGGTGTTGTACTCTGGTATTACGGTACCTTTGTACGCCTATTTT ATTTCCCCCCCCTTTTTGAAACTTAGAAGTTAATAATAAACACGCTCACTAGGTGCACTACATCCAGTAG TGTAATGAGCAAGCACTTCTGTCTYCCCCGGGAGGGATATATGGTACGCTGTGCAAACGGCGGAAATTAA TCCTACCGTTAACCGCCCACCTACTCCGAGAAGCCTAGTACCTAATTGGATTTATCAATGGAGTTGCGCT CAGCAGGTGACCCTGACCTGCCAGCTCCGGCTGATGGACCTGGGCTTTCCCCACAGGCGACTGTGGCCCA GGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGGACGCCTTGATAATGACAAGGTGGGAAGAGCCTATT GGGCTAGCTGGTTTCCTCCGGCCTCCTGAATGCGGCTAACCTTAACCCCAGAGCATATGGTAGCAACCCA GCTACTAGTATGTCATAATGCGTAAGTCTGGGATGGGACCGACTACTTTGGAGAGTCCGTGTTTCTATTG TTTCTTTAATCAATCTTATGGTGACAATTTATAGTGCCCTGAGTATTGATTGGTTGTTGCTTTTGACAAT TATTGAGACATCACATAGACATAATGGGAGCTCAAGTAAGCAGGCAAACGTCTGGTGCGCATGACACCCG GATACGGGCTGAACAGGGCGCAAACATTCATTATACTAATATCAATTATTATAGAGATGCAGCTAGCAAT GCAGCAAGCAAAATGGACTATTCCCAGGATCCGGACAAGTTCACGAAACCAGTACTTGATGCTATAACTG AACCATTACCCACGCTGAAGTCCCCTAGTGCTGAGGCATGTGGGTACAGCGACCGAGTTGCACAACTGAC AATTGGCAATTCCACTATCACTACTCAGGAAGCCGCCAATGTGGTGGTCGCATATGGACAATGGCCTGAA TATTTAGATTCGAAGGATGCAACTGCCGTGGATAAGCCCACACAGCCCGATGTAGCCTCAAATAGATTTT ACACTCTTAAGACAGTGTCTTGGGAGAAGAGTTCAACTGGCTGGTATTGGAAATTCTCGGATTGTCTGGC TTCTGTTGGATTATTTGGACAGAATGTACAGTATCATTATTTAGGCCGTTATGGGTTAGCGGTTCATGTG CAATGTAATGCTTCAAAATTTCATCAGGGCACTCTACTGGTCTTAGCAATACCAGAATGGGAGATTGGGG TGTCTAATGCTGATAGGGCATCCTTTAATCTAACAAACCCCGATAAGAACGGGCATACTATGACTGGTCA AGAAGCTTATTGCTTACATAATGGGACTAACATCCATTCTTCACTGGTATTTCCACATCAATTCATCAAT CTTAGGACAAACAATTGTGCTACGTTAGTCTTGCCCTATGTGGGAGCAACACCACTGGACACACCGATCA AGCATAATGTTTGGTCATTGGTAGTAATACCGGTGGTCCCGTTGGATTACACCACTGGTGCAACTACACA AGTGCCTATAACAATAACAATGGCTCCAATGGCGTGCGAGTTTAACGGACTGCGCAATGCCATCACCCAA GGGCTGCCAGTACTCAATACACCCGGCTCTGGGCAGTTTGTGACTACAGATAATTTCCAATCACCAAACT TGATTCCAAATTTTGATGTGACACAAGTCTTTAATAGTCCAGGTGAAATTATTAATTTACAGCAGTATGT CCAGATTGAGGGCATTATGGAAATCAATAATGTAGCAAGTGCAAATAATTTGGAGAGAATTCGCATTCCA ATATCAGTCCAGAGTGGAATTGATGAGATGTTATTTGCAATCAACTGCAACCCAGGAACAGCCCAGGAGT TTAGACGCACACCCCTGGGAGATGTGTGTAGGTATTATACACAGTGGTCAGGTAGCATACAAATTACATT TACATTTTGTGGTTCATTTATGACAACAGGAAAATTATTAATTTGCTACACCCCTCCGGGTGGTCGAGTA CCACAAAATAGAGAGGAGGCAATGCTAGGGACTAATGTGATCTGGGATTTTGGTTTACAATCCAGCGTTA CGCTGAACATACCGTGGATAAGTGGAGCCCATTTTAGAAACACTTCTGTTAATGTCGATGGTTTTGATAA CACAGGGTATGTATCTGCTTGGTTTCAAACGAACATGGTAGTTCCTCCCGATGCTCCAACGACTGCTTAT ATATTGGCTTTTACATCAGCCAAGGATGATTTCTCGATGCGCTTGTTGCGGGATACAGCAGAGATTTCGC AAGACGGATTTCTGCAAGGACCAATAGATCAAGCAATAGAAAAAGTAATCACTGATGTAGTGTCTGACAC GCGTGAGTCTAGTAGTGACTTTAGCATTGGGGCTGTTCCAGCATTGAATGCGGTGGAAACTGGAGCCACT TCGCAAGCTAGTGTTGAGTCCACCATTGAGACGCGGGCCGTGCAGAATCGTCATCGCACTTCTGAGATGA
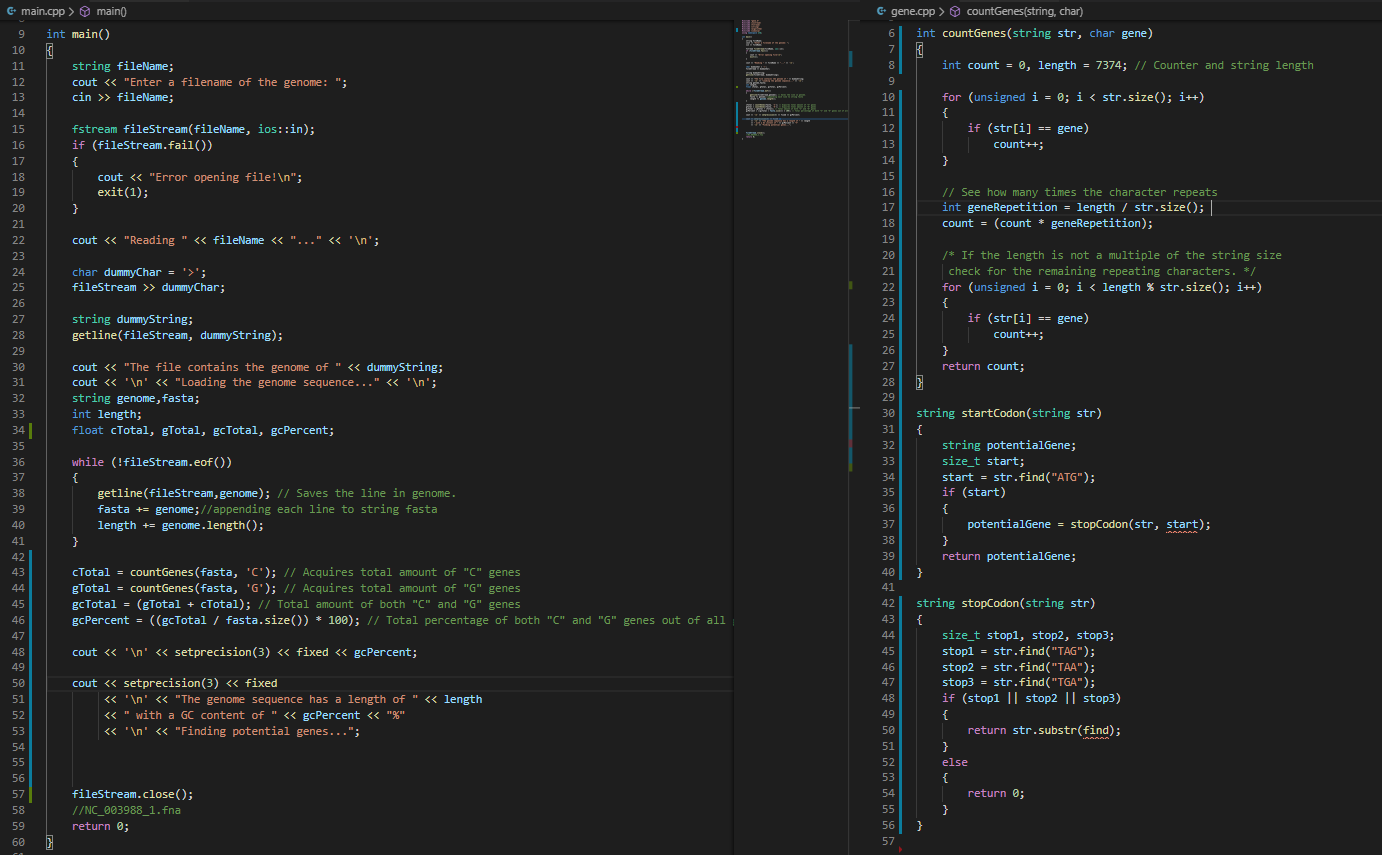
Using the string sequence, look for the start codons (start codon = ATG), look for stop codons (TAG, TAA, and TGA), and find all potential genes (defined by start and stop codons) - How to find a potential gene: MUST start with ATG and can end with TAG, TAA, or TGA Even though the sub-sequence or substring) follows the previous rule, it is NOT directly considered a potential gene. They MUST follow the rules below: The potential gene length must be divisible by 3. This is due to the fact the there MUST be an equal number of 3 characters to represent the different amino acids in the potential gene. A potential gene length of 6 is EXCLUDED. This happens when the gene basically only has the start and stop codon. In a bioinformatics research setting, this could be a configuration variable and researchers can easily change the minimum length. GTATGACAATATGA A A Frame 1 | STOP bik GLAJ GA SAAT ALGA A A Frame 2 Frame 3 GTATGACA ALALGA A A Ile [Stop Met Start codon's position ensures that this frame is chosen All the potential genes found are then written to a file and must be formatted like the sample below Arrange the genes in sorted order by the Start value which is index position where the start of the gene is located in the genome sequence string Identify the Stop value which is the index position where the gene ends in the string sequence. Also, identify the length of the potential gene Lastly, observe how Gene#2 sequence is formatted in the file. You need to print the potential gene sequence in separate lines if they exceed 72 characters. Thus, a maximum of 72 characters per line. >Gene#1 Start=144 Stop=209 Length=66 ATGAGCAAGCACTTCTGTCTYCCCCGGGAGGGATATATGGTACGCTGTGCAAACGGCGGAAATTAA >Gene#2 Start=267 Stop=458 Length=192 ATGGAGTTGCGCTCAGCAGGTGACCCTGACCTGCCAGCTCCGGCTGATGGACCTGGGCTTTCCCCACAGGCG ACTGTGGCCCAGGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGGACGCCTTGATAATGACAAGGTGGGAA GAGCCTATTGGGCTAGCTGGTTTCCTCCGGCCTCCTGAATGCGGCTAA >Gene#3 Start=313 Stop=393 Length=81 ATGGACCTGGGCTTTCCCCACAGGCGACTGTGGCCCAGGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGG ACGCCTTGA 6 gene.cpp > countGenes(string, char) 6 int countGenes(string str, char gene) e main.cpp > main( 9 int main() 10 string fileName; cout > fileName; int count = 0, length = 7374; 77 Counter and string length for (unsigned i = 0; i '; fileStream >> dummyChar; /* If the length is not a multiple of the string size check for the remaining repeating characters. */ for (unsigned i = 0; i Gene#1 Start=144 Stop=209 Length=66 ATGAGCAAGCACTTCTGTCTYCCCCGGGAGGGATATATGGTACGCTGTGCAAACGGCGGAAATTAA >Gene#2 Start=267 Stop=458 Length=192 ATGGAGTTGCGCTCAGCAGGTGACCCTGACCTGCCAGCTCCGGCTGATGGACCTGGGCTTTCCCCACAGGCG ACTGTGGCCCAGGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGGACGCCTTGATAATGACAAGGTGGGAA GAGCCTATTGGGCTAGCTGGTTTCCTCCGGCCTCCTGAATGCGGCTAA >Gene#3 Start=313 Stop=393 Length=81 ATGGACCTGGGCTTTCCCCACAGGCGACTGTGGCCCAGGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGG ACGCCTTGA 6 gene.cpp > countGenes(string, char) 6 int countGenes(string str, char gene) e main.cpp > main( 9 int main() 10 string fileName; cout > fileName; int count = 0, length = 7374; 77 Counter and string length for (unsigned i = 0; i '; fileStream >> dummyChar; /* If the length is not a multiple of the string size check for the remaining repeating characters. */ for (unsigned i = 0; i
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


