Question
Certain pathogens have what is known as a mutator phenotype associated with a high frequency of DNA mutations that enable them to change rapidly. Cryptococcus
Certain pathogens have what is known as a mutator phenotype associated with a high frequency of DNA mutations that enable them to change rapidly. Cryptococcus neoformans is a fungus that causes severe infections that are often difficult to treat. Scientists assessed whether strains of C. neoformans possess a mutator phenotype.
The scientists analyzed eleven wild, environmental strains (E1 through E11) and eleven clinical isolate strains (from patients, C1 through C11) of C. neoformans. The scientists inoculated an equal number of cells from each strain into 20 replicate liquid cultures in test tubes. After a period of growth, they removed cells from each culture and plated the cells on petri dishes containing a solid growth medium with a chemical that is typically toxic to the fungi. After an incubation period, the scientists counted the number of colonies (each colony is derived from division of a single cell) that grew in each dish and combined the data from the 20 dishes for each strain. Representative data are shown in Figure 1. The data for all environmental strains resembled the data shown for strains E1, E4, and E10. The data for all clinical strains except strains C3 and C6 resembled the data shown for strain C8.
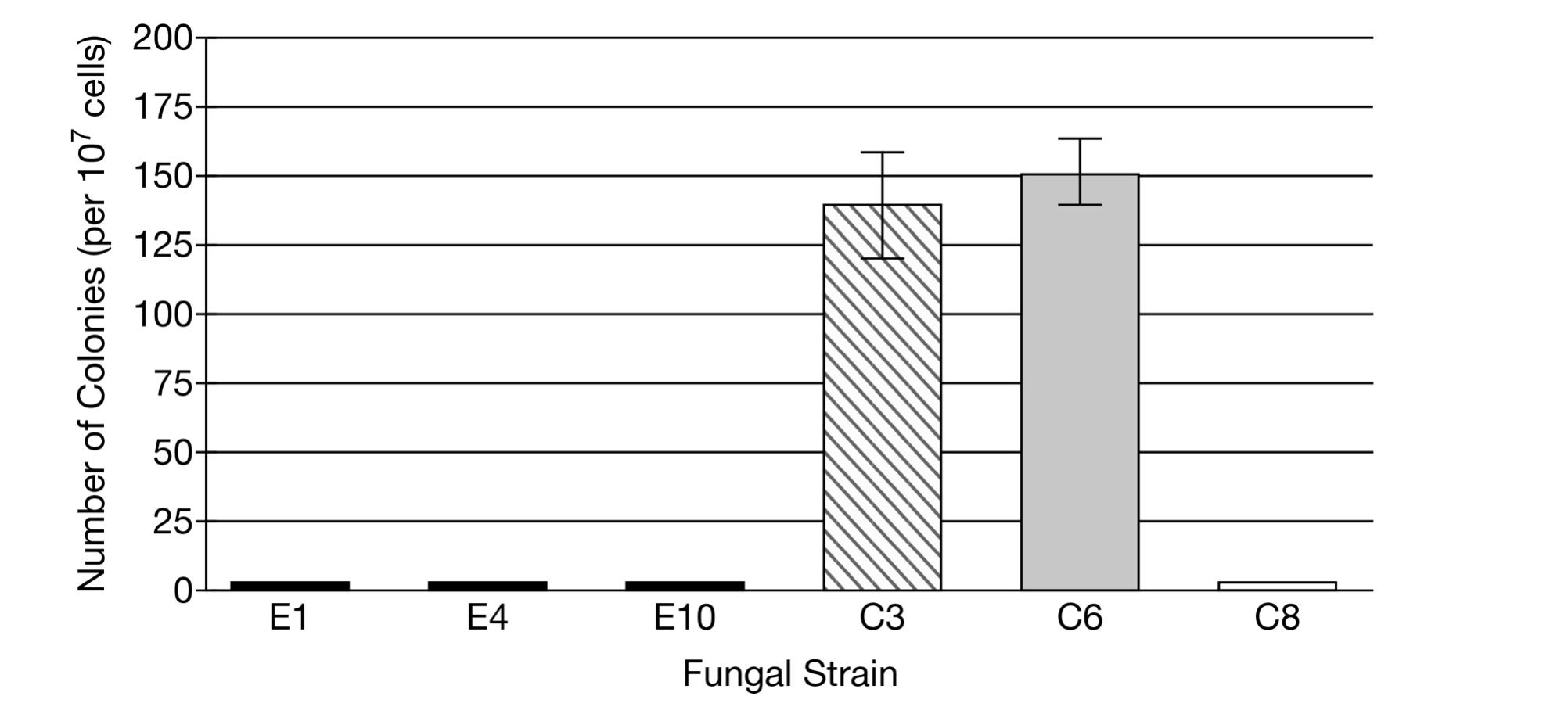
Figure 1. Number of colonies produced from C. neoformans cells grown on a medium containing a toxic chemical. Representative data from 11 environmental (E) strains and 11 clinical isolate strains (C). The error bars represent ±2SEx¯.
In a further experiment, the scientists investigated the mechanism responsible for the greater number of colonies produced by strains C3 and C6 than by the other strains. Based on DNA sequence analyses of the strains, the scientists introduced a wild-type copy of the fungal MSH2 gene, a gene encoding a repair enzyme involved in identifying nucleotides in DNA that contain incorrect bases, into cells of strains C3 and C6 and again analyzed the number of colonies that grew in the presence of the toxic chemical (Figure 2).
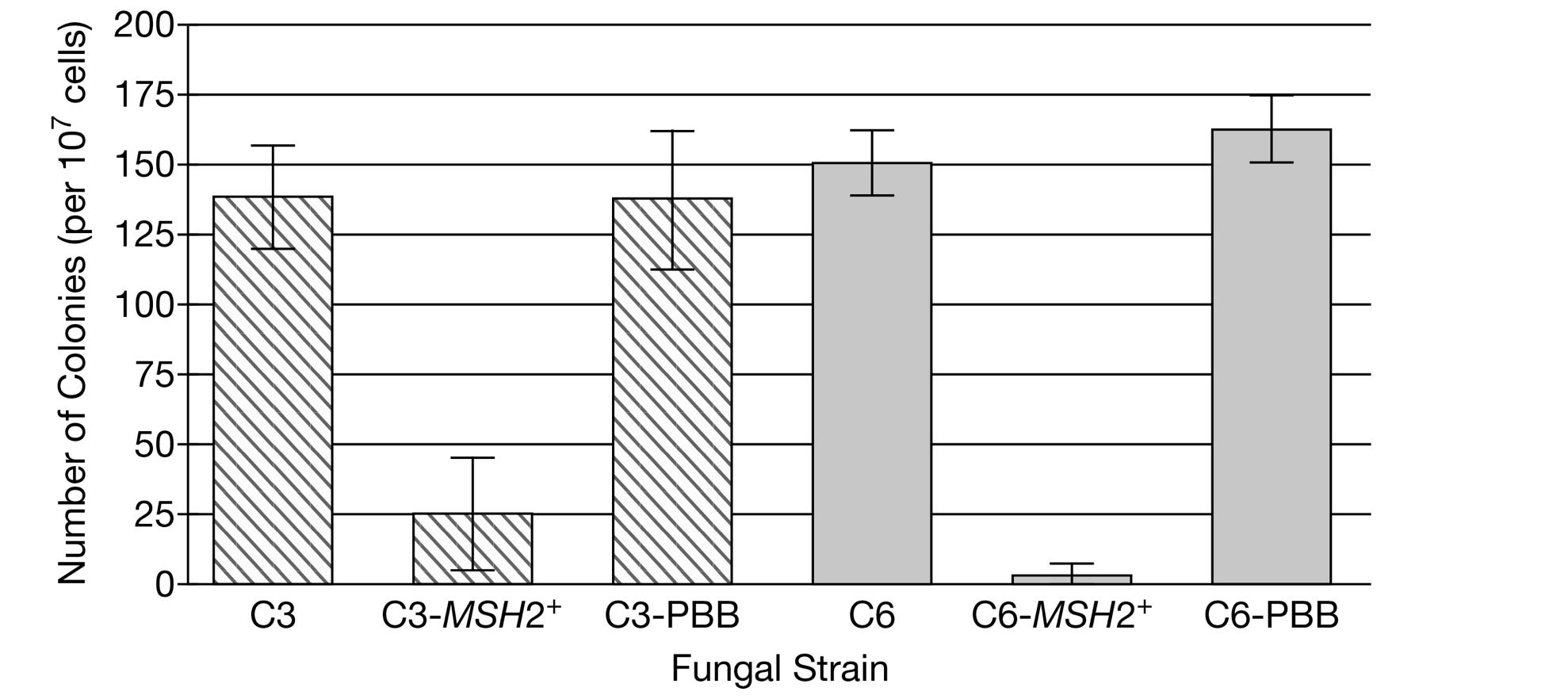
Figure 2. Number of colonies produced from C. neoformans strains C3 and C6 cells grown on a medium containing a toxic chemical. A wild-type copy of the fungal MSH2 gene was introduced into strains C3-MSH2+ and C6-MSH2+. The plasmid into which the MSH2 gene was cloned for introduction into the fungi is present in strains C3-PBB and C6-PBB. The error bars represent ±2SEx¯.
(a) Identify a dependent variable in the experiments. Identify the reasoning of the scientists when they tested the number of colonies produced by strains C3-PBB and C6-PBB. The scientists also analyzed the number of colonies produced from each of the environmental and clinical isolate strains when the strains were plated on a growth medium lacking the toxic chemical. Justify this analysis.
(b) Based on the data in Figure 1, for each strain describe the relationship between the number of colonies observed and the likely mutation rate of the strain.
(c) State the null hypothesis for the experiment whose data are graphed in Figure 2. Provide evidence to support or refute the scientists’ claim that more colonies grew in strains C3 and C6 than in the other strains because the genes for proteins that are normally targeted by the toxic chemical contain nucleotides with incorrect bases in the C3 and C6 cells. The scientists additionally determined that the C3 and C6 strains had no decrease in virulence (disease-causing ability) in comparison with the virulence of the other clinical isolate strains and concluded that these two strains have mutator phenotypes. Explain why mutator phenotypes were found only among clinical isolate strains and not among environmental strains.
200- 175- 150- 125 100- 75- 50- 25- E1 E4 E10 C3 C6 C8 Fungal Strain Number of Colonies (per 107 cells)
Step by Step Solution
3.42 Rating (149 Votes )
There are 3 Steps involved in it
Step: 1
Solution a The dependent variable is number of mutations because they inserted a gene that repairs DNA when a nucleotide is incorrectly added In the case of C3 PBB and C6 PBB they wanted to check if t...
Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


