 Python Project Help Please
Python Project Help Please
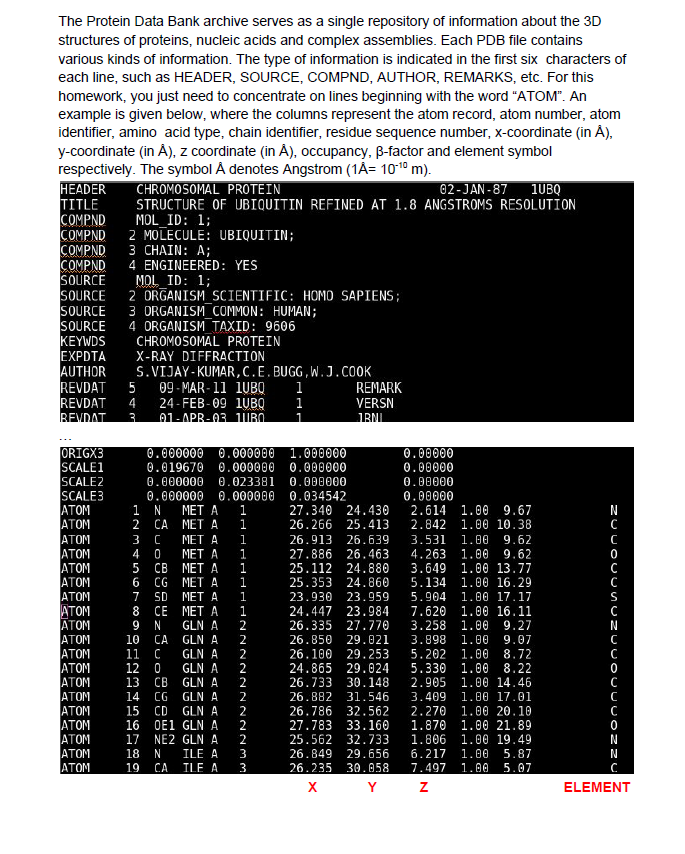
The Protein Data Bank archive serves as a single repository of information about the 3D structures of proteins, nucleic acids and complex assemblies. Each PDB file contains various kinds of information. The type of information is indicated in the first six characters of each line, such as HEADER, SOURCE, COMPND, AUTHOR, REMARKS, etc. For this homework, you just need to concentrate on lines beginning with the word "ATOM". An example is given below, where the columns represent the atom record, atom number, atom identifier, amino acid type, chain identifier, residue sequence number, x-coordinate (in A), y-coordinate (in A), z coordinate (in A), occupancy, B-factor and element symbol respectively. The symbol denotes Angstrom (1A= 10-10 m). HEADER CHROMOSOMAL PROTEIN 62-JAN-87 1UBQ TITLE STRUCTURE OF UBIQUITIN REFINED AT 1.8 ANGSTROMS RESOLUTION COMPND MOL_ID: 1; COMPND 2 MOLECULE: UBIQUITIN; COMPND 3 CHAIN: A; COMPND 4 ENGINEERED: YES SOURCE MOL ID: 1; SOURCE 2 ORGANISM SCIENTIFIC: HOMO SAPIENS; SOURCE 3 ORGANISM COMMON: HUMAN; SOURCE 4 ORGANISM TAXID: 9606 KEYWDS CHROMOSOMAL PROTEIN EXPDTA X-RAY DIFFRACTION AUTHOR S.VIJAY-KUMAR,C.E. BUGG, W.J.COOK REVDAT 09-MAR-11 IUBQ i REMARK REVDAT 4 24-FEB-09 IUBQ i VERSN REVDAT A1-APR-03 VIRO 2 1 JRM ORIGX3 SCALE1 SCALEZ SCALE3 ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM 0.000000 0.000000 0.019670 0.000000 0.000000 0.023381 0.000000 0.000000 1 N MET A 1 2 CA 1 3 C i 4 0 MET A 1 5 CB MET A 6 CG MET A 7 SD MET A 8 CE MET A 9 N GLN A 10 CA GLNA 11 C GLNA 12 0 GLNA 13 CB GLNA 14 CG GLNA 15 CD GLNA 16 OE1 GLNA 17 NE2 GLNA 18 N ILE A 19 CA ILE A 1.000000 0.000000 0.000000 0.034542 27.340 24.430 26.266 25.413 26.913 26.639 27.886 26.463 25.112 24.880 25.353 24.860 23.930 23.959 24.447 23.984 26.335 27.770 26.850 29.021 26.100 29.253 24.865 29.024 26.733 30.148 26.882 31.546 26.786 32.562 27.783 33.160 25.562 32.733 26.849 29.656 26.235 30.058 X Y 0.00000 0.00000 0.00000 0.00000 2.614 1.00 9.67 2.842 1.00 10.38 3.531 1.00 9.62 4.263 1.00 9.62 3.649 1.00 13.77 5.134 1.00 16.29 5.904 1.00 17.17 7.620 1.00 16.11 3.258 1.00 9.27 3.898 1.00 9.07 5.202 1.00 8.72 5.330 1.00 8.22 2.905 1.00 14.46 3.409 1.00 17.01 2.270 1.00 20.10 1.870 1.00 21.89 1.806 1.00 19.49 6.217 1.00 5.87 7.497 1.00 5.07 Z N C C 0 C S N WWNNNNNNNNNPPP 0 C N N N ELEMENT Implement the following functions: A. pdb_parser(pdb_filename): Receives one argument, pdb_filename. Opens the PDB file, reads all lines starting with ATOM and stores the X, Y, Z coordinates and the ELEMENT type of the atom in a list, atoms. e.g. atoms= [[27.340, 24.430, 2.614,'N'], [26.266, 25.413, 2.842, 'C']...] atoms list is of length N, number of atoms that make up the protein in the PDB file. The function should return atoms. B. center_of_mass(atoms): This function calculates the center of mass of the protein, as follows. N rc L miri Zi-mi rom is coordinates of the center of mass of the protein, r, is a list containing the [x, y, z] coordinates of the itn atom), m, is the mass of the in atom and N is the total number of atoms. m, should be obtained from the dictionary mass={'C":12.01, 'O':16.00, "H:1.008, ...} by using the element type of the itn atom as key. mass dictionary is already provided in the pdb.py template. C. shift(atoms, vec): This function translates the protein by vec and returns the updated coordinates of the protein, atomsnew. vec is a list of size 3. E.g. vec=[a, b, c), atoms=[[X, Y, Z, C']] -> atomsnew=[[X+a, y+b, Z+C, 'C']] After implementing these functions, demonstrate them as follows: i) Read the 1ubq.pdb file and parse the information. ii) Calculate the center of mass. iii) Shift the coordinates of the protein such that the molecule's center of mass is at the origin. The sample output from the program should be: Center of mass of the protein is 30.317, 28.775, 15.353 New center of mass of the protein is 0.000, 0.000, 0.000

 Python Project Help Please
Python Project Help Please





