
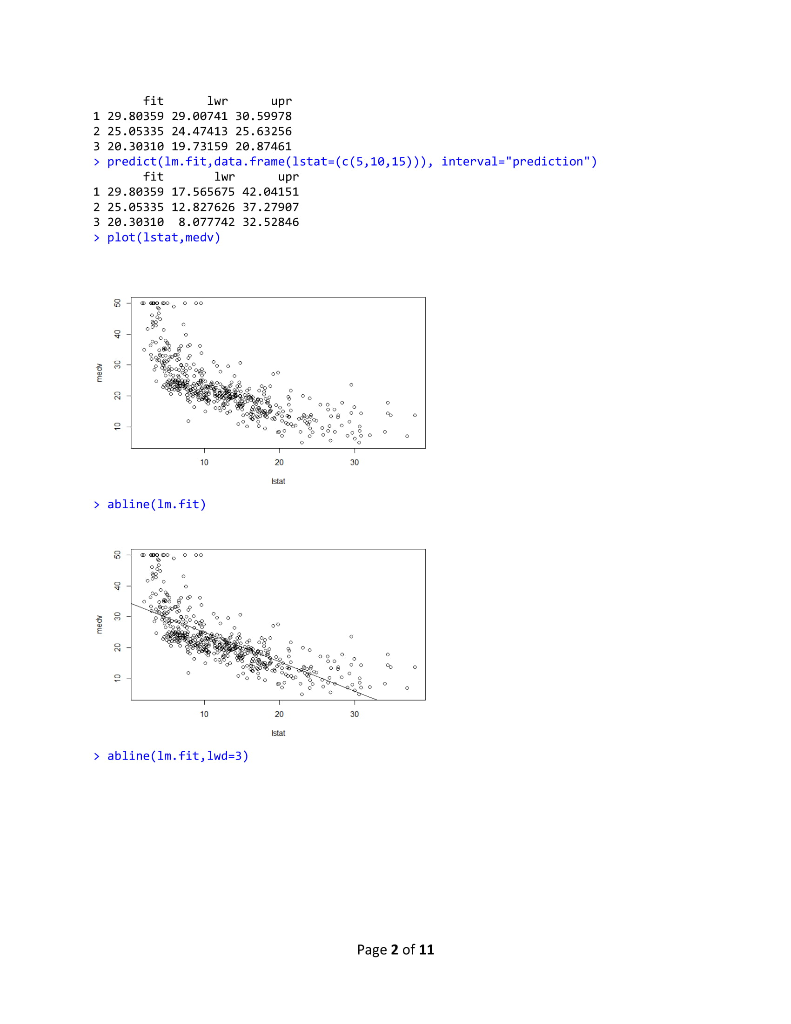
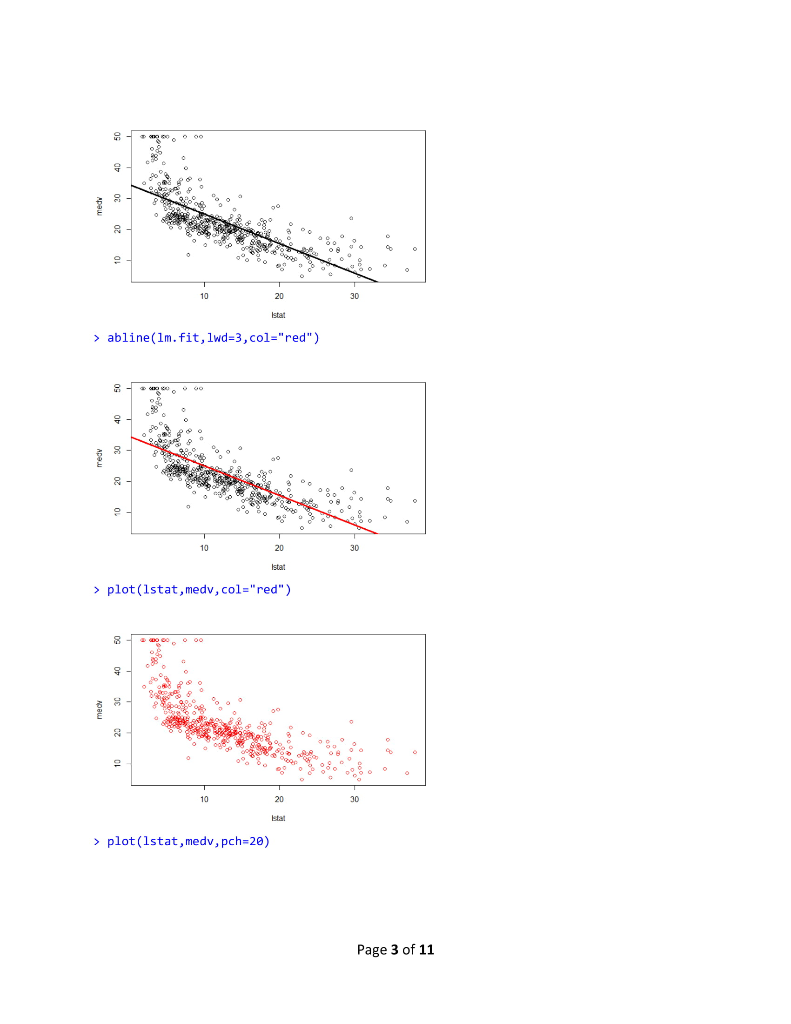
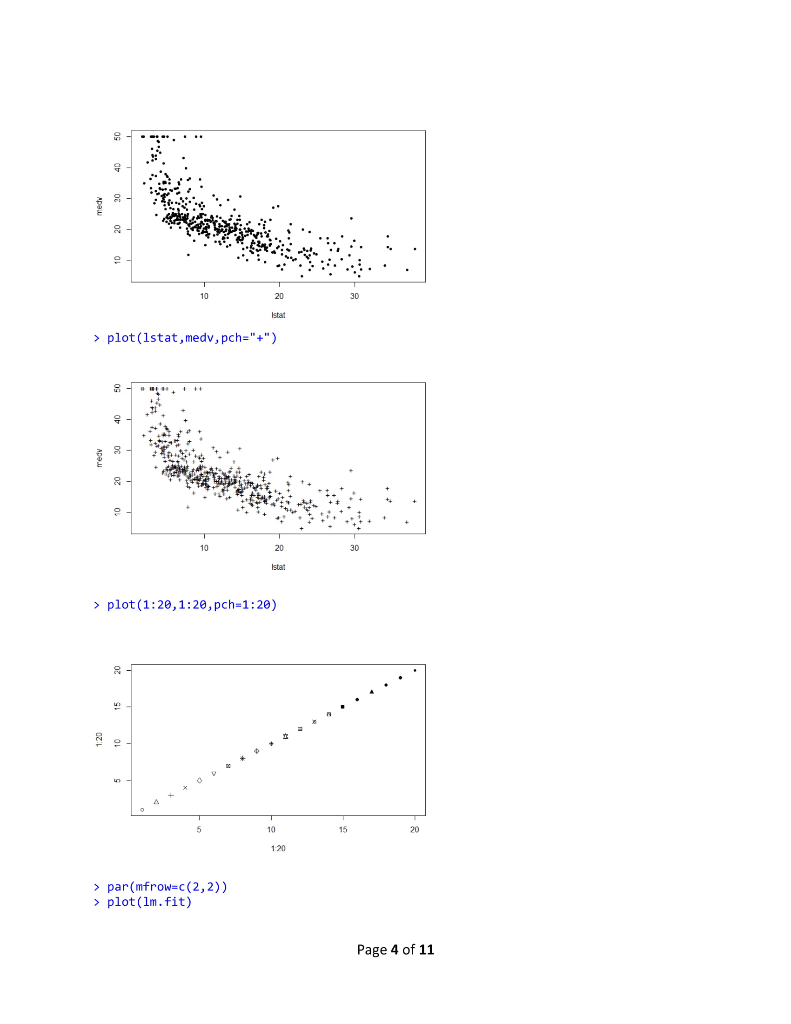
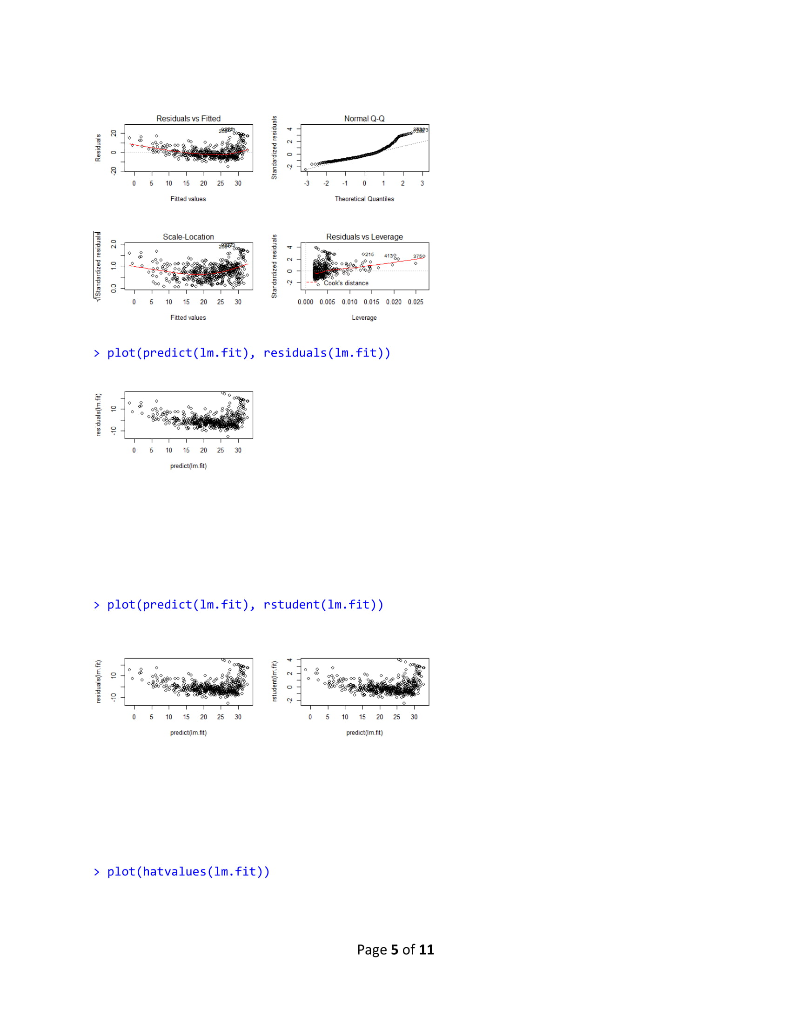
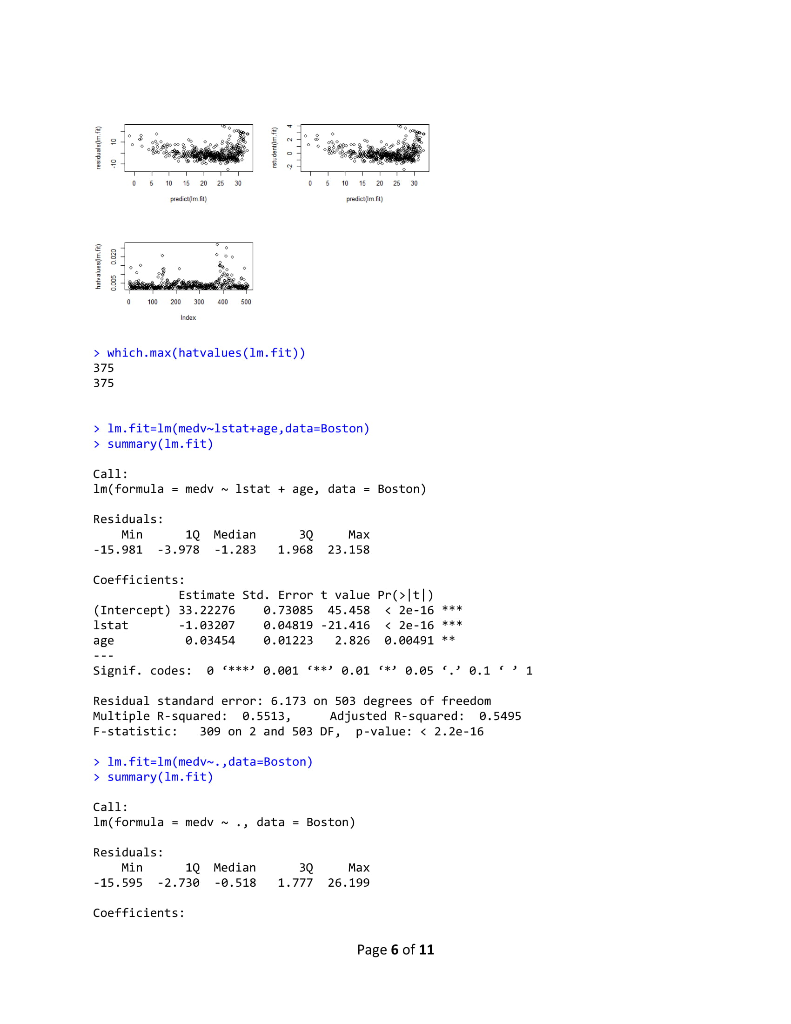
R Lab 1 Assignment > library (MASS) > library (ISLR) > fix(Boston) > names (Boston) [1] "crim" "Zn" "rad" [10] "tax" "ptratio" "black" "Istat" "medy" >1m.fit-lm(med-lstat) Error in eval (predvars, data, env) object 'medv' not found > lm.fit-1m(medv lstat, data-Boston) > attach (Boston) > 1m.fit-1m(medvlstat) > lm.fit "indus""chas" "nox" rm age "dis" Call: lm(formulamedv ~ lstat) Coefficients: (Intercept) 1stat 0.95 34.55 summary(1m.fit) Call: 1m(formula = medv ~ Istat) Residuals: Min 1Q Median 30 Max -15.168 -3.990 1.318 2.034 24.500 Coefficients: Estimate Std. Error t value Pr(>tl) (Intercept) 34.55384 .56263 61.41 names (1m.fit) [1] "coefficients" "residuals" "effects" "rank" "fitted.values" "call" [6] "assign" 6rermtst "df.residual""xlevels" [11] "terms" > coef (1m.fit) (Intercept) "model" lstat 34.5538409 -0.9500494 confint(1m.fit) 2.5 % 97.5 % (Intercept) 33.448457 35.6592247 lstat > predict (1m.fit, data.frame(1stat-(c(5,10,15))), interval-"confidence") -1.026148-0.8739505 Page 1 of 11 fit Wr upr 1 29.80359 29.00741 30.59978 2 25.05335 24.47413 25.63256 3 20.30310 19.73159 20.87461 > predict (1m.fit,data.frame (1stat-(c (5,10,15))), interval-"prediction") fit lwr upr 1 29.80359 17.565675 42.04151 2 25.05335 12.827626 37.27907 3 20.30310 8.077742 32.52846 plot(lstat,medv) 10 20 30 Istat > abline(1m.fit) 10 20 30 abline(1m.fit,lwd-3) Page 2 of 11 10 20 30 > abline(1m.fit,1wd=3,col-"red") 10 20 30 plot (1stat,medv, col-"red") 30 plot(lstat,medv, pch-20) Page 3 of 11 10 20 30 > plot (1stat,medv, pch"+") 10 20 30 > plot (1:20,1:20, pch-1:20) 10 15 20 120 > par(mfrow-c(2,2)) > plot (1m.fit) Page 4 of 11 Residuals vs Fitted Normal Q-Q 0 6 1020 2 3 2 12 3 Residuals vs Leverage Cooks dstancs 06 105 20 25 30 0000 0006 0010 0015 00 0025 Fitted values plot (predict(1m.fit), residuals (1m.fit)) 0 6 10 20 2 3 plot (predict (1m.fit), rstudent (1m.fit)) 0 5 10 5 20 25 30 0 5 10 15 0 25 30 pred ct)m.t predict)im ft) > plot (hatvalues (1m.fit)) Page 5 of 11 0 6 1020 2 3 0 10 20 25 3 0 100 200 3400 500 which.max(hatvalues (1m.fit)) 375 375 >1m.fit-1m(medvlstat+age, data-Boston) > summary(lm.fit) Call 1m(formulamedv lstat age, data -Boston) Residuals: -15.981 3.978 -1.283 1.968 23.158 Coefficients: Min 1Q Median 3Q Max Estimate Std. Error t value Pr(>lt|) (Intercept) 33.22276 0.73085 45.458 2e-16 lstat age 1.03207 0.04819-21.416 lm.fit-1m(medv~,data-Boston) > summary (lm.fit) Call 1m(formula-medv ~ ., data-Boston) Residuals 15.595 2.730 0.518 1.777 26.199 Coefficients: Min 1Q Median 3Q Max Page 6 of 11 R Lab 1 Assignment > library (MASS) > library (ISLR) > fix(Boston) > names (Boston) [1] "crim" "Zn" "rad" [10] "tax" "ptratio" "black" "Istat" "medy" >1m.fit-lm(med-lstat) Error in eval (predvars, data, env) object 'medv' not found > lm.fit-1m(medv lstat, data-Boston) > attach (Boston) > 1m.fit-1m(medvlstat) > lm.fit "indus""chas" "nox" rm age "dis" Call: lm(formulamedv ~ lstat) Coefficients: (Intercept) 1stat 0.95 34.55 summary(1m.fit) Call: 1m(formula = medv ~ Istat) Residuals: Min 1Q Median 30 Max -15.168 -3.990 1.318 2.034 24.500 Coefficients: Estimate Std. Error t value Pr(>tl) (Intercept) 34.55384 .56263 61.41 names (1m.fit) [1] "coefficients" "residuals" "effects" "rank" "fitted.values" "call" [6] "assign" 6rermtst "df.residual""xlevels" [11] "terms" > coef (1m.fit) (Intercept) "model" lstat 34.5538409 -0.9500494 confint(1m.fit) 2.5 % 97.5 % (Intercept) 33.448457 35.6592247 lstat > predict (1m.fit, data.frame(1stat-(c(5,10,15))), interval-"confidence") -1.026148-0.8739505 Page 1 of 11 fit Wr upr 1 29.80359 29.00741 30.59978 2 25.05335 24.47413 25.63256 3 20.30310 19.73159 20.87461 > predict (1m.fit,data.frame (1stat-(c (5,10,15))), interval-"prediction") fit lwr upr 1 29.80359 17.565675 42.04151 2 25.05335 12.827626 37.27907 3 20.30310 8.077742 32.52846 plot(lstat,medv) 10 20 30 Istat > abline(1m.fit) 10 20 30 abline(1m.fit,lwd-3) Page 2 of 11 10 20 30 > abline(1m.fit,1wd=3,col-"red") 10 20 30 plot (1stat,medv, col-"red") 30 plot(lstat,medv, pch-20) Page 3 of 11 10 20 30 > plot (1stat,medv, pch"+") 10 20 30 > plot (1:20,1:20, pch-1:20) 10 15 20 120 > par(mfrow-c(2,2)) > plot (1m.fit) Page 4 of 11 Residuals vs Fitted Normal Q-Q 0 6 1020 2 3 2 12 3 Residuals vs Leverage Cooks dstancs 06 105 20 25 30 0000 0006 0010 0015 00 0025 Fitted values plot (predict(1m.fit), residuals (1m.fit)) 0 6 10 20 2 3 plot (predict (1m.fit), rstudent (1m.fit)) 0 5 10 5 20 25 30 0 5 10 15 0 25 30 pred ct)m.t predict)im ft) > plot (hatvalues (1m.fit)) Page 5 of 11 0 6 1020 2 3 0 10 20 25 3 0 100 200 3400 500 which.max(hatvalues (1m.fit)) 375 375 >1m.fit-1m(medvlstat+age, data-Boston) > summary(lm.fit) Call 1m(formulamedv lstat age, data -Boston) Residuals: -15.981 3.978 -1.283 1.968 23.158 Coefficients: Min 1Q Median 3Q Max Estimate Std. Error t value Pr(>lt|) (Intercept) 33.22276 0.73085 45.458 2e-16 lstat age 1.03207 0.04819-21.416 lm.fit-1m(medv~,data-Boston) > summary (lm.fit) Call 1m(formula-medv ~ ., data-Boston) Residuals 15.595 2.730 0.518 1.777 26.199 Coefficients: Min 1Q Median 3Q Max Page 6 of 11












