Question
THE GENETICS FILE IS BELOW. Copy and Paste into Notepad++ and name it Genetics.PY and use this file to import into the program file. GENE

THE GENETICS FILE IS BELOW. Copy and Paste into Notepad++ and name it Genetics.PY and use this file to import into the program file.
GENE = """ >ENA|BAE76126|BAE76126.1 Escherichia coli str. K-12 substr. W3110 beta-D-galactosidase ATGACCATGATTACGGATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCT GGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGC GAAGAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGCGC TTTGCCTGGTTTCCGGCACCAGAAGCGGTGCCGGAAAGCTGGCTGGAGTGCGATCTTCCT GAGGCCGATACTGTCGTCGTCCCCTCAAACTGGCAGATGCACGGTTACGATGCGCCCATC TACACCAACGTGACCTATCCCATTACGGTCAATCCGCCGTTTGTTCCCACGGAGAATCCG ACGGGTTGTTACTCGCTCACATTTAATGTTGATGAAAGCTGGCTACAGGAAGGCCAGACG CGAATTATTTTTGATGGCGTTAACTCGGCGTTTCATCTGTGGTGCAACGGGCGCTGGGTC GGTTACGGCCAGGACAGTCGTTTGCCGTCTGAATTTGACCTGAGCGCATTTTTACGCGCC GGAGAAAACCGCCTCGCGGTGATGGTGCTGCGCTGGAGTGACGGCAGTTATCTGGAAGAT CAGGATATGTGGCGGATGAGCGGCATTTTCCGTGACGTCTCGTTGCTGCATAAACCGACT ACACAAATCAGCGATTTCCATGTTGCCACTCGCTTTAATGATGATTTCAGCCGCGCTGTA CTGGAGGCTGAAGTTCAGATGTGCGGCGAGTTGCGTGACTACCTACGGGTAACAGTTTCT TTATGGCAGGGTGAAACGCAGGTCGCCAGCGGCACCGCGCCTTTCGGCGGTGAAATTATC GATGAGCGTGGTGGTTATGCCGATCGCGTCACACTACGTCTGAACGTCGAAAACCCGAAA CTGTGGAGCGCCGAAATCCCGAATCTCTATCGTGCGGTGGTTGAACTGCACACCGCCGAC GGCACGCTGATTGAAGCAGAAGCCTGCGATGTCGGTTTCCGCGAGGTGCGGATTGAAAAT GGTCTGCTGCTGCTGAACGGCAAGCCGTTGCTGATTCGAGGCGTTAACCGTCACGAGCAT CATCCTCTGCATGGTCAGGTCATGGATGAGCAGACGATGGTGCAGGATATCCTGCTGATG AAGCAGAACAACTTTAACGCCGTGCGCTGTTCGCATTATCCGAACCATCCGCTGTGGTAC ACGCTGTGCGACCGCTACGGCCTGTATGTGGTGGATGAAGCCAATATTGAAACCCACGGC ATGGTGCCAATGAATCGTCTGACCGATGATCCGCGCTGGCTACCGGCGATGAGCGAACGC GTAACGCGAATGGTGCAGCGCGATCGTAATCACCCGAGTGTGATCATCTGGTCGCTGGGG AATGAATCAGGCCACGGCGCTAATCACGACGCGCTGTATCGCTGGATCAAATCTGTCGAT CCTTCCCGCCCGGTGCAGTATGAAGGCGGCGGAGCCGACACCACGGCCACCGATATTATT TGCCCGATGTACGCGCGCGTGGATGAAGACCAGCCCTTCCCGGCTGTGCCGAAATGGTCC ATCAAAAAATGGCTTTCGCTACCTGGAGAGACGCGCCCGCTGATCCTTTGCGAATACGCC CACGCGATGGGTAACAGTCTTGGCGGTTTCGCTAAATACTGGCAGGCGTTTCGTCAGTAT CCCCGTTTACAGGGCGGCTTCGTCTGGGACTGGGTGGATCAGTCGCTGATTAAATATGAT GAAAACGGCAACCCGTGGTCGGCTTACGGCGGTGATTTTGGCGATACGCCGAACGATCGC CAGTTCTGTATGAACGGTCTGGTCTTTGCCGACCGCACGCCGCATCCAGCGCTGACGGAA GCAAAACACCAGCAGCAGTTTTTCCAGTTCCGTTTATCCGGGCAAACCATCGAAGTGACC AGCGAATACCTGTTCCGTCATAGCGATAACGAGCTCCTGCACTGGATGGTGGCGCTGGAT GGTAAGCCGCTGGCAAGCGGTGAAGTGCCTCTGGATGTCGCTCCACAAGGTAAACAGTTG ATTGAACTGCCTGAACTACCGCAGCCGGAGAGCGCCGGGCAACTCTGGCTCACAGTACGC GTAGTGCAACCGAACGCGACCGCATGGTCAGAAGCCGGGCACATCAGCGCCTGGCAGCAG TGGCGTCTGGCGGAAAACCTCAGTGTGACGCTCCCCGCCGCGTCCCACGCCATCCCGCAT CTGACCACCAGCGAAATGGATTTTTGCATCGAGCTGGGTAATAAGCGTTGGCAATTTAAC CGCCAGTCAGGCTTTCTTTCACAGATGTGGATTGGCGATAAAAAACAACTGCTGACGCCG CTGCGCGATCAGTTCACCCGTGCACCGCTGGATAACGACATTGGCGTAAGTGAAGCGACC CGCATTGACCCTAACGCCTGGGTCGAACGCTGGAAGGCGGCGGGCCATTACCAGGCCGAA GCAGCGTTGTTGCAGTGCACGGCAGATACACTTGCTGATGCGGTGCTGATTACGACCGCT CACGCGTGGCAGCATCAGGGGAAAACCTTATTTATCAGCCGGAAAACCTACCGGATTGAT GGTAGTGGTCAAATGGCGATTACCGTTGATGTTGAAGTGGCGAGCGATACACCGCATCCG GCGCGGATTGGCCTGAACTGCCAGCTGGCGCAGGTAGCAGAGCGGGTAAACTGGCTCGGA TTAGGGCCGCAAGAAAACTATCCCGACCGCCTTACTGCCGCCTGTTTTGACCGCTGGGAT CTGCCATTGTCAGACATGTATACCCCGTACGTCTTCCCGAGCGAAAACGGTCTGCGCTGC GGGACGCGCGAATTGAATTATGGCCCACACCAGTGGCGCGGCGACTTCCAGTTCAACATC AGCCGCTACAGTCAACAGCAACTGATGGAAACCAGCCATCGCCATCTGCTGCACGCGGAA GAAGGCACATGGCTGAATATCGACGGTTTCCATATGGGGATTGGTGGCGACGACTCCTGG AGCCCGTCAGTATCGGCGGAATTCCAGCTGAGCGCCGGTCGCTACCATTACCAGTTGGTC TGGTGTCAAAAATAA """
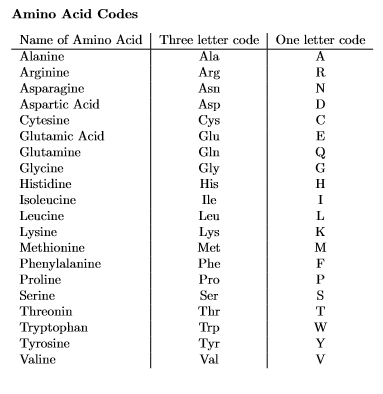
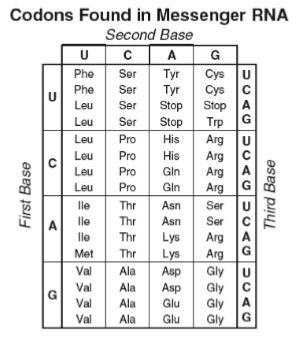
Write the program in Python. This program is used to understand string processing 1. Import the GENETICS variable from the genetics.py module 2. Change the DEOXYRIBONUCLEIC acid sequence to an RIBONUCLEIC acid sequence.A DEOXYRIBONUCLEIC sequence is composed of four bases: adenine (A). cytosine (C). guanine (G) and thymine (T). An RIBONUCLEIC sequence contains uracil (U) instead of thymine (T), but the other bases are the same 3. 4. Use list/dictionaries for the program to create the table below 4. Translate the RIBONUCLEIC acid RNA sequence to an amino acid sequence. Start by splitting the RIBONUCLEIC acid sequence into segments of length three, called codons, and then translating each codon into an amino acid 5. In the program remove the line >ENAIBAE761261BAE76126.1 Escherichia coli str. K-12 substr. W3110 beta-D-galactosidase" from the file, but do not remove it from the Genetics.py file 6. Print out the amino acid sequence as a string of one letter amino acid codes. If a given codon does not have a corresponding amino acid. then print an hashtag
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


