You work at a medical testing facility, conducting genetics tests and familial genetic screening to investigate more unusual cases of human disease. Usually your work
You work at a medical testing facility, conducting genetics tests and familial genetic screening to investigate more unusual cases of human disease. Usually your work involves known inherited genetic diseases. In this case a whole family have been referred to you for immediate genetic analyses.
A male (aged 35) has been suffering with stomach cramps and severe, watery diarrhoea. He has had the symptoms for over 48 hours and attended the accident and emergency department at the hospital. He was immediately treated for dehydration and once the hospital discovered he had recently returned from a safari trip to sub-Saharan Africa, a series of tests were undertaken. A range of diagnostic genetic tests using a sample of his blood have been conducted using polymerase chain reaction (PCR) and DNA sequencing. All the tests come back as normal apart from one, in the battery of viral detection tests, an unusual result arose (see figure one below).

Once the result for the test were confirmed, the hospital immediately contacted the patients’ family and called them in for testing. The couple had been on holiday together, although the wife was not suffering any symptoms except a mild stomach discomfort, which she attributed to travelling abroad. Her result for the same viral detection test showed she was also infected. The couple have a three-year-old son, who did not travel with them and is not experiencing any new symptoms since their return, but the parents informed the hospital that they were concerned about his health as recently he had been rather unwell, with bowel discomfort and poor weight gain.
There can be several reasons for a differential response to infection. All family members were immediately quarantined and further biochemical and genetic tests (alongside treatment) were conducted.
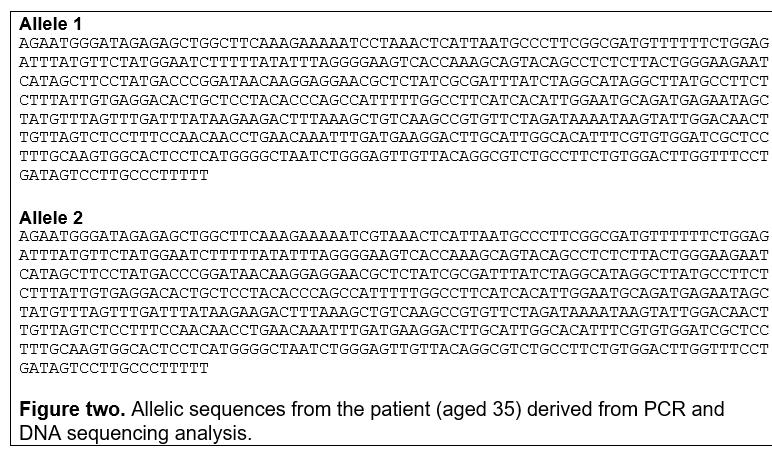
Results of one of the genetic tests are provided below:



Please complete all the following tasks using the information provided.
Task one
Identify the sequence in Figure 1 by BLAST analysis. What infection has the patient contracted? Ensure that you demonstrate use of bioinformatics tools by including screen shots (with legends) of your analyses and an appropriate explanation of the data.
Task two
Identify the sequences in Figures 2-5 by BLAST analysis. Provide the following information: sequence name, gene name, database accession number, chromosomal location, size (bp) and exon count of the (full) gene. Ensure that you demonstrate use of bioinformatics tools by including screen shots of your analyses and appropriate explanations of the data.
Hint: You only need to provide an exemplar screen shot of one BLAST analysis, but all data should be discussed in the explanation.
Task three
As you are interested in the potential genetic differences so you further analyse the sequence data. Use an online alignment tool to align all the nucleotide sequences from Figures 2-4 alongside the database-derived sequence in figure 5. Provide a picture of your alignment, then identify and fully explain (using appropriate literature) any differences between the sequences.
Task four
It is important to verify if any sequence abnormalities that occur at a nucleotide level will affect the peptide sequence. Use an online translation tool and translate all the nucleotide sequences from figures 2-5 into amino acid sequences. Then align amino acid sequences using an online alignment tool. Provide a picture of your alignment and identify and explain any differences between the sequences.
Task five
You are interested in any structural effects on the protein caused by the mutations found. Use the ProtCalc software (hosted by JustBio) to determine if the mutations affect the molecular weight and pI of the peptide sequences (just the parts you have translated). Determine whether the protein function is likely to be affected.
Task Six
You are interested in the results you have obtained and think that they may be important in explaining the differences in the responses of each family member to the infection. To find this out, undertake a literature search to identify any links between the gene you have identified, the mutations and the infection. Is there any scientific literature, which links the gene/protein and associated mutations with susceptibility to the disease? Investigate the evidence and illustrate concepts with figures where necessary.
Task Seven
Find the accession page for sequence NM_000492.3, which encodes the full mRNA for your gene of interest.
Using this sequence, design primers for amplification of the coding sequence into the pPAL7 vector for expression as an N terminal fusion protein in E.coli.
Details of pPAL 7 vector
http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_5867.pdf
Your strategy should cover all aspects of the cloning process and should be justified with scientific explanations throughout. The following aspects will need to be covered and you should ensure to include appropriate screen shots or other information and diagrams to evidence your strategy. Ensure you show full evidence of your analysis and design, giving annotated screenshots and web addresses where appropriate.
- Outline of cloning strategy to produce the recombinant plasmid construct with appropriate justification at all points.
- Design appropriate primers to allow the DNA to be cloned into the vector to enable the expression of the protein as an N terminal fusion protein with the Profinity exact tag which will help you to be able to purify the expressed protein. Provide justification for your choice of primers and highlight all the important aspects of the primers that you have designed and give the PCR cycling conditions that would be appropriate for amplification.
- Details of controls to be included at various points throughout this experimental process to allow you to determine the causes of any problems etc.
GATACACATAATAGAATTAAGGATGAATTATGATTAAATTAAAATTTGG TGTTTTTTTTACAGTTTTACTATCTTCAGCATATGCACATGGAACACCT CAAAATATTACTGATTTGTGTGCAGAATACCACAACACACAAATACATA CGCTAAATGATAAGATATTGTCGTATACAGAATCTCTAGCTGGAAACAG AGAGATGGCTATCATTACTTTTAAGAATGGTGCAACTTTTCAAGTAGAA GTACCAGGTAGTCAACATATAGATTCACAAAAAAAAGCGATTGAAAGGA TGAAGGATACCCTGAGGATTGCATATCTTACTGAAGCTAAAGTCGAAAA GTTATGTGTATGGAATAATAAAACGCCTCATGCGATTGCCGCAATTAGT ATGGCAAATTAAGATATAAAAAAGCCCA Figure One. Sequencing data from PCR detection test #451a. Male aged 35 yrs.
Step by Step Solution
3.42 Rating (168 Votes )
There are 3 Steps involved in it
Step: 1
An Introduction to Cloning A researchers guide to cloning DNA We offer a number of proven technologi...
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


