An article in Analytical Biochemistry [Application of Central Composite Design for DNA Hybridization Onto Magnetic Microparticles, (2009,
Question:
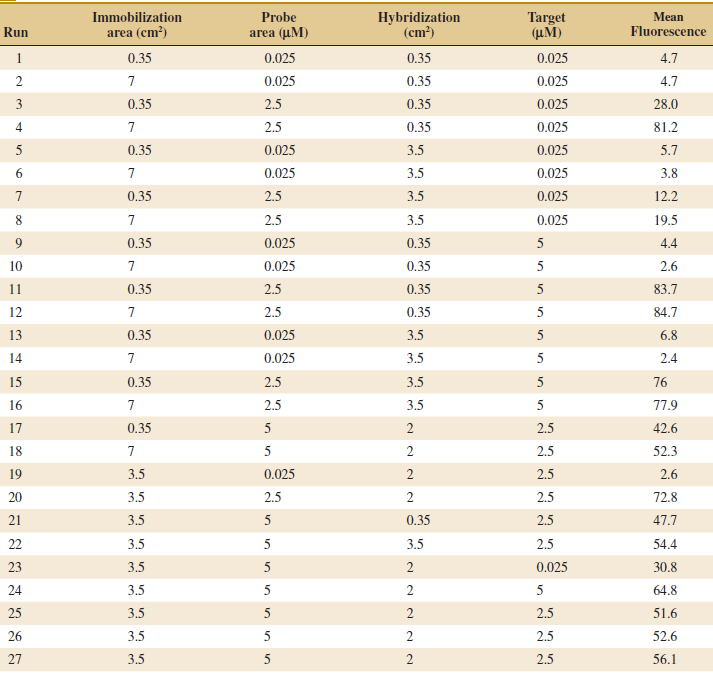
(a) What type of design is used?
(b) Fit a second-order response surface model to the data.
(c) Does a residual analysis indicate any problems?
Fantastic news! We've Found the answer you've been seeking!
Step by Step Answer:
Related Book For 

Applied Statistics And Probability For Engineers
ISBN: 9781118539712
6th Edition
Authors: Douglas C. Montgomery, George C. Runger
Question Posted:





