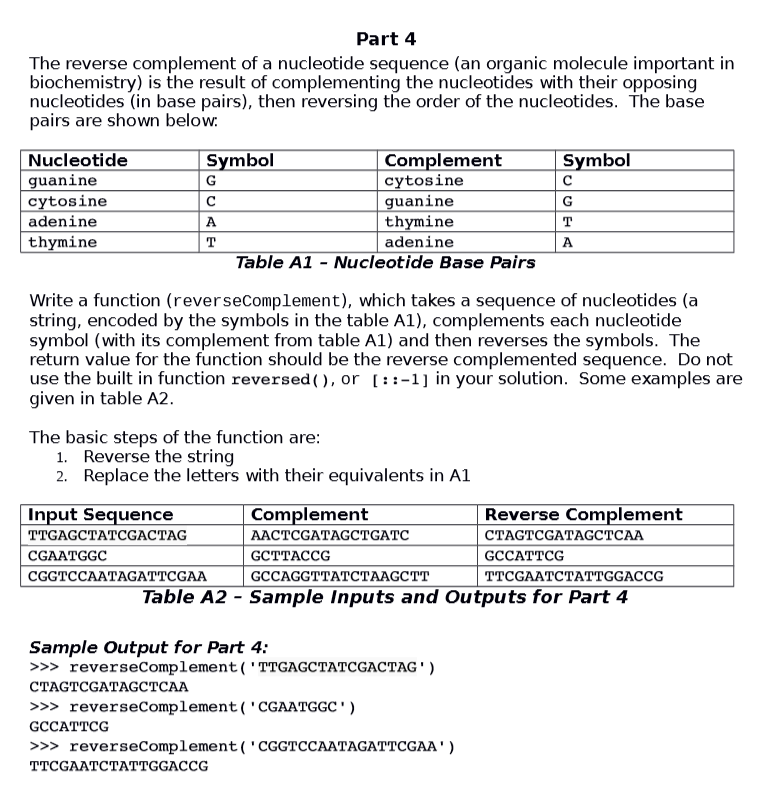
Question
Here is the first part of my code: import random def dnaMolecule(length): return ''.join(random.choice('GCAT') for x in range(length)) var = dnaMolecule(19) #choose length of GCAT
Here is the first part of my code:
import random
def dnaMolecule(length): return ''.join(random.choice('GCAT') for x in range(length))
var = dnaMolecule(19) #choose length of GCAT print ('The molecules are:', var) #prints dnaMolecule(x - 1) characters
gcount = var.count('G') #counts g and c content seperately ccount = var.count('C') gc = gcount + ccount
total = len(var)
gcContent = (gc / total) * 100 #divides the amount of gc by # of characters in string print('The concentration of GC is:',"%.2f" % gcContent, '%') #%.2f rounds to 2 decimal places
What is the proper way for me to take the reverse complement of my random string in this code?
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


