Question
I have a data ( 172, 168, 175, 173, 179, 175 ) And I have three prior distribution of , N(169,5) , N(172,5), N(169,10) How
I have a data ( 172, 168, 175, 173, 179, 175 )
And I have three prior distribution of 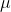 , N(169,5) , N(172,5), N(169,10)
, N(169,5) , N(172,5), N(169,10)
How can I make Graph of posterior of these by BernGrid.R code?
BernGrid = function( Theta , pTheta , Data , plotType=c("Points","Bars")[2] ,
showCentTend=c("Mean","Mode","None")[3] ,
showHDI=c(TRUE,FALSE)[2] , HDImass=0.95 ,
showpD=c(TRUE,FALSE)[2] , nToPlot=length(Theta) ) {
# Theta is vector of values between 0 and 1.
# pTheta is prior probability mass at each value of Theta
# Data is vector of 0's and 1's.
# Check for input errors:
if ( any( Theta > 1 | Theta
stop("Theta values must be between 0 and 1")
}
if ( any( pTheta
stop("pTheta values must be non-negative")
}
if ( !isTRUE(all.equal( sum(pTheta) , 1.0 )) ) {
stop("pTheta values must sum to 1.0")
}
if ( !all( Data == 1 | Data == 0 ) ) {
stop("Data values must be 0 or 1")
}
# Create summary values of Data
z = sum( Data ) # number of 1's in Data
N = length( Data )
# Compute the Bernoulli likelihood at each value of Theta:
pDataGivenTheta = Theta^z * (1-Theta)^(N-z)
# Compute the evidence and the posterior via Bayes' rule:
pData = sum( pDataGivenTheta * pTheta )
pThetaGivenData = pDataGivenTheta * pTheta / pData
# Plot the results.
layout( matrix( c( 1,2,3 ) ,nrow=3 ,ncol=1 ,byrow=FALSE ) ) # 3x1 panels
par( mar=c(3,3,1,0) , mgp=c(2,0.7,0) , mai=c(0.5,0.5,0.3,0.1) ) # margins
cexAxis = 1.33
cexLab = 1.75
# convert plotType to notation used by plot:
if ( plotType=="Points" ) { plotType="p" }
if ( plotType=="Bars" ) { plotType="h" }
dotsize = 5 # how big to make the plotted dots
barsize = 5 # how wide to make the bar lines
# If the comb has a zillion teeth, it's too many to plot, so plot only a
# thinned out subset of the teeth.
nteeth = length(Theta)
if ( nteeth > nToPlot ) {
thinIdx = round( seq( 1, nteeth , length=nteeth ) )
} else {
thinIdx = 1:nteeth
}
# Plot the prior.
yLim = c(0,1.1*max(c(pTheta,pThetaGivenData)))
plot( Theta[thinIdx] , pTheta[thinIdx] , type=plotType ,
pch="." , cex=dotsize , lwd=barsize ,
xlim=c(0,1) , ylim=yLim , cex.axis=cexAxis ,
xlab=bquote(theta) , ylab=bquote(p(theta)) , cex.lab=cexLab ,
main="Prior" , cex.main=1.5 , col="skyblue" )
if ( showCentTend != "None" ) {
if ( showCentTend == "Mean" ) {
meanTheta = sum( Theta * pTheta )
if ( meanTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , yLim[2] ,
bquote( "mean=" * .(signif(meanTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
if ( showCentTend == "Mode" ) {
modeTheta = Theta[ which.max( pTheta ) ]
if ( modeTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , yLim[2] ,
bquote( "mode=" * .(signif(modeTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
}
# Mark the highest density interval. HDI points are not thinned in the plot.
if ( showHDI ) {
HDIinfo = HDIofGrid( pTheta , credMass=HDImass )
points( Theta[ HDIinfo$indices ] ,
rep( HDIinfo$height , length( HDIinfo$indices ) ) ,
pch="-" , cex=1.0 )
text( mean( Theta[ HDIinfo$indices ] ) , HDIinfo$height ,
bquote( .(100*signif(HDIinfo$mass,3)) * "% HDI" ) ,
adj=c(0.5,-1.5) , cex=1.5 )
# Mark the left and right ends of the waterline.
# Find indices at ends of sub-intervals:
inLim = HDIinfo$indices[1] # first point
for ( idx in 2:(length(HDIinfo$indices)-1) ) {
if ( ( HDIinfo$indices[idx] != HDIinfo$indices[idx-1]+1 ) | # jumps on left, OR
( HDIinfo$indices[idx] != HDIinfo$indices[idx+1]-1 ) ) { # jumps on right
inLim = c(inLim,HDIinfo$indices[idx]) # include idx
}
}
inLim = c(inLim,HDIinfo$indices[length(HDIinfo$indices)]) # last point
# Mark vertical lines at ends of sub-intervals:
for ( idx in inLim ) {
lines( c(Theta[idx],Theta[idx]) , c(-0.5,HDIinfo$height) , type="l" , lty=2 ,
lwd=1.5 )
text( Theta[idx] , HDIinfo$height , bquote(.(round(Theta[idx],3))) ,
adj=c(0.5,-0.1) , cex=1.2 )
}
}
# Plot the likelihood: p(Data|Theta)
plot( Theta[thinIdx] , pDataGivenTheta[thinIdx] , type=plotType ,
pch="." , cex=dotsize , lwd=barsize ,
xlim=c(0,1) , ylim=c(0,1.1*max(pDataGivenTheta)) , cex.axis=cexAxis ,
xlab=bquote(theta) , ylab=bquote( "p(D|" * theta * ")" ) , cex.lab=cexLab ,
main="Likelihood" , cex.main=1.5 , col="skyblue" )
if ( z > .5*N ) { textx = 0 ; textadj = c(0,1) }
else { textx = 1 ; textadj = c(1,1) }
text( textx ,1.0*max(pDataGivenTheta) ,cex=2.0
,bquote( "Data: z=" * .(z) * ",N=" * .(N) ) ,adj=textadj )
if ( showCentTend != "None" ) {
if ( showCentTend == "Mean" ) {
meanTheta = sum( Theta * pDataGivenTheta )
if ( meanTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , 0.7*max(pDataGivenTheta) ,
bquote( "mean=" * .(signif(meanTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
if ( showCentTend == "Mode" ) {
modeTheta = Theta[ which.max( pDataGivenTheta ) ]
if ( modeTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , 0.7*max(pDataGivenTheta) ,
bquote( "mode=" * .(signif(modeTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
}
# Plot the posterior.
yLim = c(0,1.1*max(c(pTheta,pThetaGivenData)))
plot( Theta[thinIdx] , pThetaGivenData[thinIdx] , type=plotType ,
pch="." , cex=dotsize , lwd=barsize ,
xlim=c(0,1) , ylim=yLim , cex.axis=cexAxis ,
xlab=bquote(theta) , ylab=bquote( "p(" * theta * "|D)" ) , cex.lab=cexLab ,
main="Posterior" , cex.main=1.5 , col="skyblue" )
if ( showCentTend != "None" ) {
if ( showCentTend == "Mean" ) {
meanTheta = sum( Theta * pThetaGivenData )
if ( meanTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , yLim[2] ,
bquote( "mean=" * .(signif(meanTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
if ( showCentTend == "Mode" ) {
modeTheta = Theta[ which.max( pThetaGivenData ) ]
if ( modeTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , yLim[2] ,
bquote( "mode=" * .(signif(modeTheta,3)) ) ,
cex=2.0 , adj=textadj )
}
}
# Plot marginal likelihood pData:
if ( showpD ) {
meanTheta = sum( Theta * pThetaGivenData )
if ( meanTheta > .5 ) {
textx = 0 ; textadj = c(0,1)
} else {
textx = 1 ; textadj = c(1,1)
}
text( textx , 0.75*max(pThetaGivenData) , cex=2.0 ,
bquote( "p(D)=" * .(signif(pData,3)) ) ,adj=textadj )
}
# Mark the highest density interval. HDI points are not thinned in the plot.
if ( showHDI ) {
HDIinfo = HDIofGrid( pThetaGivenData , credMass=HDImass )
points( Theta[ HDIinfo$indices ] ,
rep( HDIinfo$height , length( HDIinfo$indices ) ) ,
pch="-" , cex=1.0 )
text( mean( Theta[ HDIinfo$indices ] ) , HDIinfo$height ,
bquote( .(100*signif(HDIinfo$mass,3)) * "% HDI" ) ,
adj=c(0.5,-1.5) , cex=1.5 )
# Mark the left and right ends of the waterline.
# Find indices at ends of sub-intervals:
inLim = HDIinfo$indices[1] # first point
for ( idx in 2:(length(HDIinfo$indices)-1) ) {
if ( ( HDIinfo$indices[idx] != HDIinfo$indices[idx-1]+1 ) | # jumps on left, OR
( HDIinfo$indices[idx] != HDIinfo$indices[idx+1]-1 ) ) { # jumps on right
inLim = c(inLim,HDIinfo$indices[idx]) # include idx
}
}
inLim = c(inLim,HDIinfo$indices[length(HDIinfo$indices)]) # last point
# Mark vertical lines at ends of sub-intervals:
for ( idx in inLim ) {
lines( c(Theta[idx],Theta[idx]) , c(-0.5,HDIinfo$height) , type="l" , lty=2 ,
lwd=1.5 )
text( Theta[idx] , HDIinfo$height , bquote(.(round(Theta[idx],3))) ,
adj=c(0.5,-0.1) , cex=1.2 )
}
}
return( pThetaGivenData )
} # end of function
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


