Question
I have the code as well edit it and give me in different way. void FASTAreadset_LL:: read_unsorted(char * fn){ // cout > str; //cout >

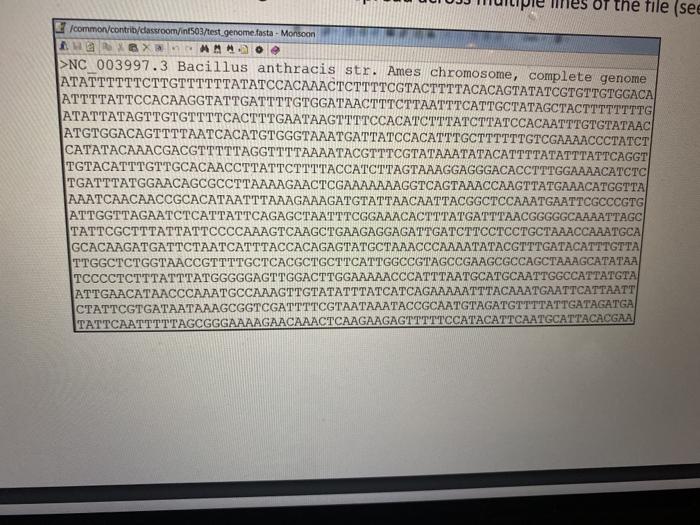
I have the code as well edit it and give me in different way.
void FASTAreadset_LL:: read_unsorted(char * fn){ // cout > str; //cout > full_file[len]; len++; } //cout =50){ str = s.substr(0,50); s.erase(0,1); // output = strlen(full_file)) break; } if(j >= strlen(full_file)) break; count++; output Problem #2 (of 2): Basic search Read in the Bacillus anthracis genome (/common/contrib/classroom/inf503/test_genome.fasta). Read in the sequence read fragments used in problem #1 - you must use the FASTAreadset_LL created in the previous problem to store your sequence reads. A. Break down the genome sequence into all 50-character long fragments contained within (shifting start location by one base each time). Store these fragments in an array or linked list data structure (your choice). How many 50 character fragments did you observe in the genome? B. Iterate through all 50-mers found in the genome, using the search function you developed in 1D to query the read set. How many genome 50-mer fragments were found in your read set? How long it take to complete the entire search process (all 50-mers)? Note that the problem 18 may take a LONG time to complete. If you feel that you will not be able to complete the execution of the program in time to submit the assignment, estimate the total amount of time and the number of found 50-mers by running 1000, 10000, 100000 searches and estimating (extrapolating) the outcome from those, OT the file (see /common/contrib/classroom/int503/test genome.fasta - Monsoon Ia X AMM. >NC_003997.3 Bacillus anthracis str. Ames chromosome, complete genome ATATTTTTTCTTGTTTTTTATATCCACAAACTCTTTTCGTACTTTTACACAGTATATCGTGTTGTGGACA ATTTTATTCCACAAGGTATTGATTTTGTGGATAACTTTCTTAATITCATTGCTATAGCTACTTTTTTTTG ATATTATAGTTGTGTTTTCACTTTGAATAAGTTTTCCACATCTTTATOTTATCCACAATTTGTGTATAAC ATGTGGACAGTTTTAATCACATGTGGGTAAATGATTATCCACATTTGCTTTTTTGTCGAAAACCCTATCT CATATACAAACGACGTTTTTAGGTTTTAAAATACGTTTCGTATAAATATACATTTTATATTTATTCAGGT TGTACATTTGTTGCACAACCTTATTCTTTTACCATCTTAGTAAAGGAGGGACACCTTTGGAAAACATCTC TGATTTATGGAACAGCGCCTTAAAAGAACTCGAAAAAAAGGTCAGTAAACCAAGTTATGAAACATGUTTA AAATCAACAACCGCACATAATTTAAAGAAAGATGTATTAACAATTACGGCTCCAAATGAATTCGCCCGTG ATTGGTTAGAATCTCATTATTCAGAGCTAATTTCGGAAACACTITATGATTTAACGGGGGCAAAATTAGC TATTCGCTTTATTATTCCCCAAAGTCAAGCTGAAGAGGAGATTGATCTTCCTCCTGCTAAACCAAATGCA GCACAAGATGATTCTAATCATTTACCACAGAGTATGCTAAACCCAAAATATACGTTTGATACATTTGTTA TTGGCTCTGGTAACCGTTTTGCTCACGCTGCTTCATTGGCCGTAGCCGAAGCGCCAGCTAAAGCATATAA TCCCCTCTTTATTTATGGGGGAGTTGGACTTGGAAAAACCCATTTAATGCATGCAATTGGCCATTATGTA ATTGAACATAACCCAAATGCCAAAGTTGTATATTTATCATCAGAAAAATTTACAAATGAATTCATTAATT CTATTCGTGATAATAAAGCGGTCGATTTTCGTAATAAATACCGCAATGTAGATGTTTTATTGATAGATGA TATTCAATTTTTAGCGGGAAAAGAACAAACTCAAGAAGAGTTTTTCCATACATTCAATGCATTACACGAA Aa March 3, 2021 at 10:34 PM Please help me in writing the code for those 2 questions. Problem #2 (of 2): Basic search Read in the Bacillus anthracis genome (/common/contrib/classroom/inf503/test_genome.fasta). Read in the sequence read fragments used in problem #1 - you must use the FASTAreadset_LL created in the previous problem to store your sequence reads. A. Break down the genome sequence into all 50-character long fragments contained within (shifting start location by one base each time). Store these fragments in an array or linked list data structure (your choice). How many 50 character fragments did you observe in the genome? B. Iterate through all 50-mers found in the genome, using the search function you developed in 1D to query the read set. How many genome 50-mer fragments were found in your read set? How long it take to complete the entire search process (all 50-mers)? Note that the problem 18 may take a LONG time to complete. If you feel that you will not be able to complete the execution of the program in time to submit the assignment, estimate the total amount of time and the number of found 50-mers by running 1000, 10000, 100000 searches and estimating (extrapolating) the outcome from those, OT the file (see /common/contrib/classroom/int503/test genome.fasta - Monsoon Ia X AMM. >NC_003997.3 Bacillus anthracis str. Ames chromosome, complete genome ATATTTTTTCTTGTTTTTTATATCCACAAACTCTTTTCGTACTTTTACACAGTATATCGTGTTGTGGACA ATTTTATTCCACAAGGTATTGATTTTGTGGATAACTTTCTTAATITCATTGCTATAGCTACTTTTTTTTG ATATTATAGTTGTGTTTTCACTTTGAATAAGTTTTCCACATCTTTATOTTATCCACAATTTGTGTATAAC ATGTGGACAGTTTTAATCACATGTGGGTAAATGATTATCCACATTTGCTTTTTTGTCGAAAACCCTATCT CATATACAAACGACGTTTTTAGGTTTTAAAATACGTTTCGTATAAATATACATTTTATATTTATTCAGGT TGTACATTTGTTGCACAACCTTATTCTTTTACCATCTTAGTAAAGGAGGGACACCTTTGGAAAACATCTC TGATTTATGGAACAGCGCCTTAAAAGAACTCGAAAAAAAGGTCAGTAAACCAAGTTATGAAACATGUTTA AAATCAACAACCGCACATAATTTAAAGAAAGATGTATTAACAATTACGGCTCCAAATGAATTCGCCCGTG ATTGGTTAGAATCTCATTATTCAGAGCTAATTTCGGAAACACTITATGATTTAACGGGGGCAAAATTAGC TATTCGCTTTATTATTCCCCAAAGTCAAGCTGAAGAGGAGATTGATCTTCCTCCTGCTAAACCAAATGCA GCACAAGATGATTCTAATCATTTACCACAGAGTATGCTAAACCCAAAATATACGTTTGATACATTTGTTA TTGGCTCTGGTAACCGTTTTGCTCACGCTGCTTCATTGGCCGTAGCCGAAGCGCCAGCTAAAGCATATAA TCCCCTCTTTATTTATGGGGGAGTTGGACTTGGAAAAACCCATTTAATGCATGCAATTGGCCATTATGTA ATTGAACATAACCCAAATGCCAAAGTTGTATATTTATCATCAGAAAAATTTACAAATGAATTCATTAATT CTATTCGTGATAATAAAGCGGTCGATTTTCGTAATAAATACCGCAATGTAGATGTTTTATTGATAGATGA TATTCAATTTTTAGCGGGAAAAGAACAAACTCAAGAAGAGTTTTTCCATACATTCAATGCATTACACGAA Aa March 3, 2021 at 10:34 PM Please help me in writing the code for those 2 questions
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


