Question
I just need answers for all TODO. I do not need any explanation or any other details. Just only answer for all TODO. TODO 6
I just need answers for all TODO. I do not need any explanation or any other details. Just only answer for all TODO.
TODO 6
To complete the TODO finish the below AddPolynomialFeatures class. The below class works by computing the polynomial feature transform from the passed data and creating/returning a new DataFrame containing both old and new polynomial features. The class takes in an argument called degree which determines the degree of the polynomial that will be computed.
Compute the polynomial for the current feature column_name based on the current degree d. To do so, you need to index data X at the current column using the name of the column stored in column_name. Once you index the current feature's/column's data, you need to raise data to the current power determined by d. Store the output into degree_feat.
Hint: To raise data to a given power you can use the ** syntax (ref) or you can use NumPy's power() function (docs).
Append the degree_name to the column_names list using its append() method.
Append the degree_feat to the features list using its append() method.
Concatenate our old and new polynomial features into a new DataFrame using Panda's concat() function (docs).
Hint: Be sure to specify the correct axis for the axis argument! We want to concatenate our features so that they are side-by-side. Meaning, they are stacked column-wise!
class AddPolynomialFeatures(BaseEstimator, TransformerMixin): def __init__(self, degree): self.feature_names = None self.degree = degree def fit(self, X: pd.DataFrame) -> None: return self def transform(self, X: pd.DataFrame) -> pd.DataFrame: # Return if the degree is only 1 as no transform # needs to be computed. if self.degree # TODO 6.1 degree_feat = # Determines the column name for our # new DataFrame which contains the Polynomial # and original features. if d > 1: degree_name = column_name+f"^{d}" else: degree_name = column_name # TODO 6.2 # TODO 6.3 # TODO 6.4 poly_X = # Set column names for our new polynomial DataFrame poly_X.columns = column_names # Set the new feature names self.feature_names = poly_X.columns return poly_X def get_feature_names_out(self, name=None) -> pd.Series: return self.feature_names
TODO 7
Complete the TODO by finishing the feature_pipeline() function.
Take note that this function is a little different from our target_pipeline() function. Notice, we have to use the Sklearn Pipeline class in conjunction with our DataFrameColumnTransformer class. This is because we want the AddBias and Standardization classes to apply to our entire data while we want the AddPolynomialFeatures and OneHotEncoding classes to only apply to certain columns/features.
Recall, we use our DataFrameColumnTransformer class to apply certain data cleaning and transformations to certain columns. We use Sklearn's Pipeline class to apply certain data cleaning and transformations to ALL the columns.
To begin, we need to construct our column transformer's data cleaning and transformation stages. These stages will consist of AddPolynomialFeatures, OneHotEncoding for 'month', and OneHotEncoding for 'day'. We separate the 'month' and 'day' one-hot encodings in case we want to drop one of these two features. When encoding them both at the same time we could break the ColumnTransformer class if we dropped either 'month' or 'day' as it wouldn't be able to find the feature.Create the polynomial transform stage by appending a tuple to our list col_trans_stages. The tuple should be defined as follows :
Element 1 should be set to the string 'poly_transform'.
Element 2 should be set an instance of our AddPolynomialFeatures class and pass the poly_degree argument to determine the degree of the polynomial.
Element 3 should be the names of the features we want the polynomial transform to be applied to. This is given by the poly_col_names argument.
Next, create the one-hot encoding stage for 'month' by appending a tuple to our list col_trans_stages. The tuple should be defined as follows
Element 1 should be set to the string 'one_hot_month'.
Element 2 should be set an instance of our OneHotEncoding class.
Element 3 should be set to a list containing the 'month' feature name.
Lastly, create the one-hot encoding stage for 'day' by appending a tuple to our list col_trans_stages. The tuple should be defined as follows
Element 1 should be set to the string 'one_hot_day'.
Element 2 should be set an instance of our OneHotEncoding class.
Element 3 should be set to a list containing the 'day' feature name.
def feature_pipeline( X_trn: pd.DataFrame, X_vld: pd.DataFrame, X_tst: pd.DataFrame, poly_degree: int = 1, poly_col_names: List[str] = []) -> List[pd.DataFrame]: """ Creates column transformers and pipelines to apply data clean and transfornations to the input features of our data. Args: X_trn: Train data. X_vld: Validation data. X_tst: Test data. poly_degree: Polynomial degrees which is passed to AddPolynomialFeatures. poly_col_names: Name of features to apply polynomial transform to. """ col_trans_stages = []
if poly_degree > 1: col_trans_stages.append( # TODO 7.1.A )
if 'month' in X_trn.columns:
col_trans_stages.append( # TODO 7.1.B ) if 'day' in X_trn.columns: col_trans_stages.append( # TODO 7.1.C ) # Create DataFrameColumnTransformer that takes in col_trans_stages feature_col_trans = DataFrameColumnTransformer(col_trans_stages) # Create a Pipeline that calls our feature_col_trans and # other transformation classes that apply to ALL data. feature_pipe = Pipeline([ ('col_transformer', feature_col_trans), ('scaler', Standardization()), ('bias', AddBias()), ]) # Fit and transform data X_trn_clean = feature_pipe.fit_transform(X_trn) X_vld_clean = feature_pipe.transform(X_vld) X_tst_clean = feature_pipe.transform(X_tst) return X_trn_clean, X_vld_clean, X_tst_clean
TODO 8
def reshape_labels(y: np.ndarray) -> np.ndarray: if len(y.shape) == 1: y = y.reshape(-1, 1) return y
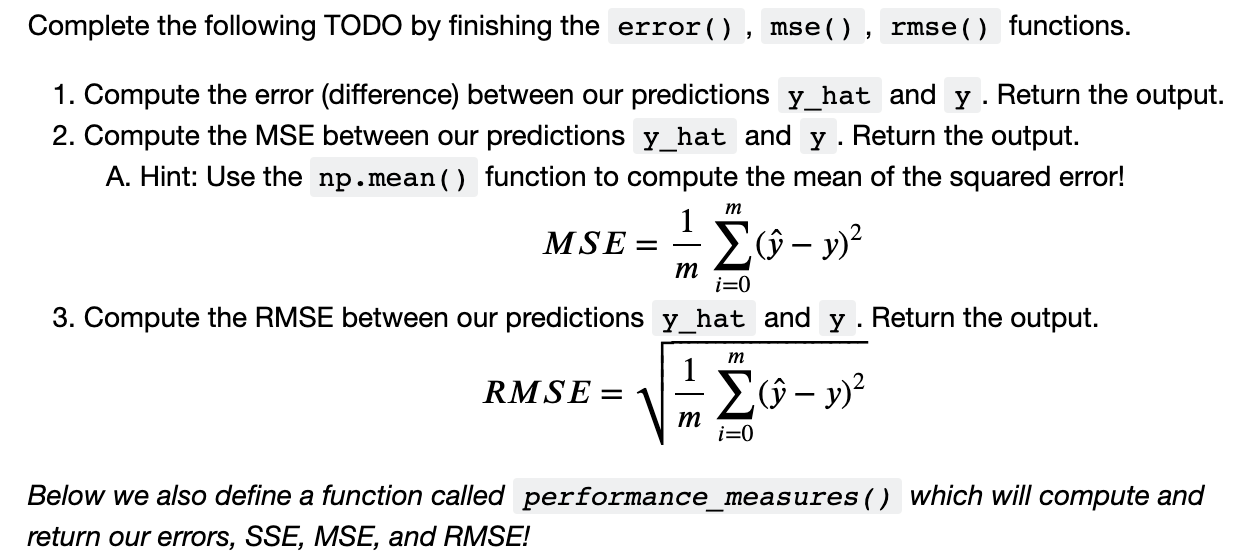
def error(y: np.ndarray, y_hat: np.ndarray) -> np.ndarray: y = reshape_labels(y) y_hat = reshape_labels(y_hat) # TODO 8.1 pass # Replace this line with your code
def mse(y: np.ndarray, y_hat: np.ndarray) -> np.ndarray: y = reshape_labels(y) y_hat = reshape_labels(y_hat) # TODO 8.2 pass # Replace this line with your code
def rmse(y: np.ndarray, y_hat: np.ndarray) -> np.ndarray: y = reshape_labels(y) y_hat = reshape_labels(y_hat) # TODO 8.3 pass # Replace this line with your code
def performance_measures(y: np.ndarray, y_hat: np.ndarray) -> Tuple[np.ndarray]: y = reshape_labels(y) y_hat = reshape_labels(y_hat) err = error(y=y, y_hat=y_hat) sse = np.sum(err**2) mse_ = mse(y=y, y_hat=y_hat) rmse_ = rmse(y=y, y_hat=y_hat) return err, sse, mse_, rmse_
TODO 12
Complete the TODO by getting our data, training the OrdinaryLeastSquares class and making predictions for our training and validation data.
Call the data_prep() function to get our cleaned and transformed data. Store the output into data. Make sure to pass forestfire_df and the arguments corresponding to the following descriptions:
Return all data as NumPy arrays.
Drop the features 'day', 'ISI', 'DC', 'RH', and 'FFMC' from the data using the drop_features keyword argument.
Create an instance of the OrdinaryLeastSquares and pass the =10=10 to the model using the lamb keyword argument. Store the output into ols.
# TODO 12.1 data = X_trn, y_trn, X_vld, y_vld, _, _, feature_names = data
# TODO 12.2 ols =
ols.fit(X_trn, y_trn)
y_hat_trn = ols.predict(X_trn)
_, trn_sse, trn_mse, trn_rmse = analyze( y=y_trn, y_hat=y_hat_trn, title="Training Predictions Log Transform", dataset="Training", xlabel="Data Sample Index", ylabel="Predicted Log Area" )
todo_check([ (isinstance(X_trn, np.ndarray), 'X_trn is not of type np.ndarray'), (np.isclose(trn_rmse, 1.34096, rtol=.01), "trn_rmse value is possibly incorrect!"), (np.all(np.isclose(ols.w[:3].flatten(), [ 1.1418, -0.03522, -0.0148 ], rtol=0.01)), 'ols.w weights possibly contain incorrect values!') ])
Complete the following TODO by finishing the , , functions. 1. Compute the error (difference) between our predictions and . Return the output. 2. Compute the MSE between our predictions and y. Return the output. A. Hint: Use the function to compute the mean of the squared error! MSE=m1i=0m(y^y)2 3. Compute the RMSE between our predictions y_hat and y. Return the output. RMSE=m1i=0m(y^y)2 Below we also define a function called performance_measures () which will compute and return our errors, SSE, MSE, and RMSEStep by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


