Implement the code written below into the Gene.py module(1st, 2nd, and 3rd photos). Then use the test code to check your work.(4th photo)
def mutate_seq(seq, pos, base): new_seq = seq[:pos] + base + seq[pos+1:] return new_seq
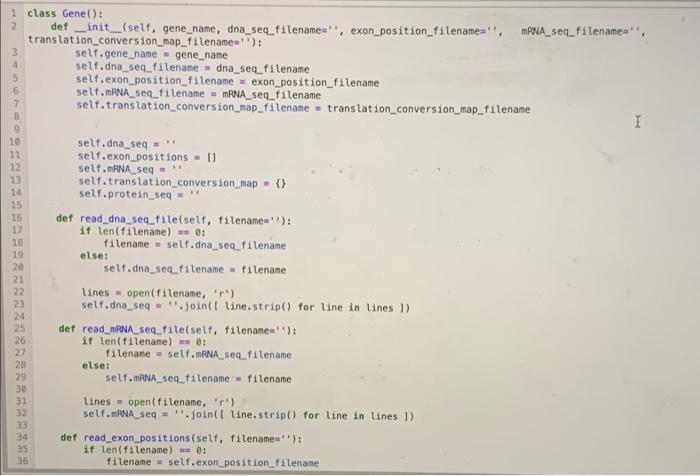
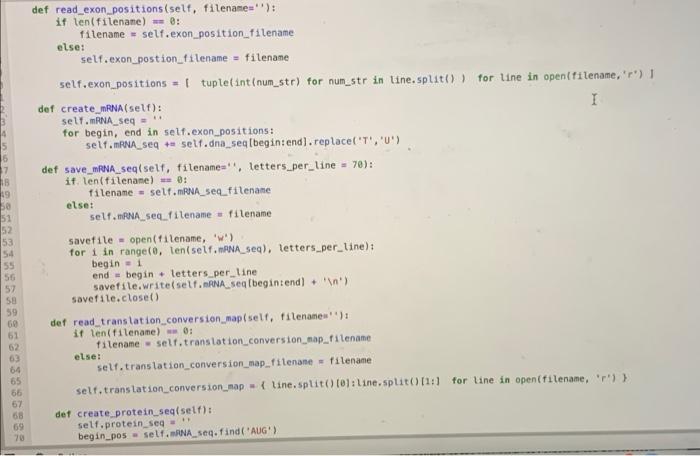
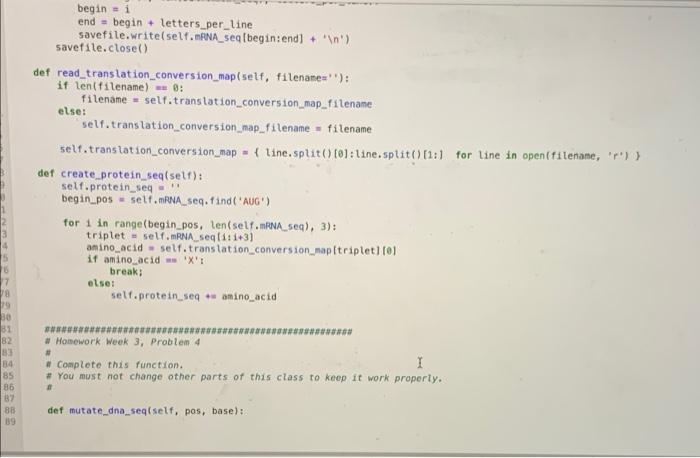
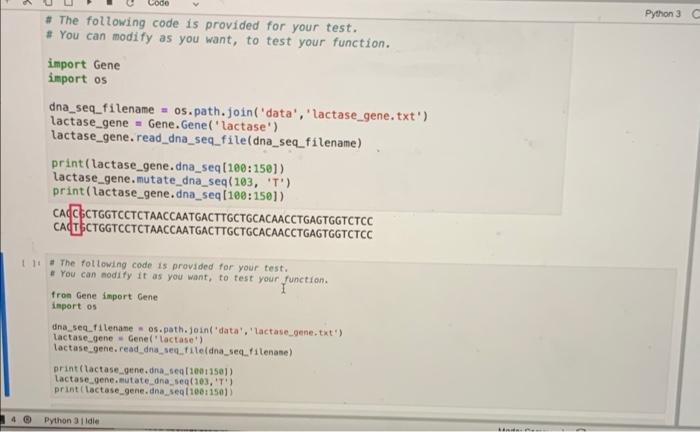
10 1 class Gene(): 2 def __init_(self, gene_name, dna_seq_filename=", exon_position_filename=' translation_conversion_map_filename='); mRNA_seq_filename, self.gene_name gene_name selt.dna seg_filename = dna_seq_filename self.exon_position_filename = exon_position_filename self.mRNA seq_filename = mRNA seq_filename self.translation_conversion_map_filename = translation_conversion_map_filename I self.dna_seq=1 21 self.exon_positions - 11 12 self.mRNA_seg - 13 self. translation conversion_map) 14 self.protein_seq=1 15 16 det read_dna_seq_file(self, filename'); 17 it len(filename) - 0: 18 filename = self.dna_seq_filename else: 20 self.dna_seq. Filename - filenane 21 22 lines open(filename, 'r) 23 self.dno_seg - ".joint line.strip() for line in lines) 24 25 def read_mRNA seq.fileself, filename): 26 if len(filename) -- : 27 Filename = self.mRNA_seq_filename 28 else: 29 self. RNA seq_filename = filename Lines = open(filename, '') self. RNA_seq= ".joint line.strip) for line in lines 1) 33 def read_exon_positions(self, filename'): 35 if len(filename) : 36 filename = self.exon_position filename 19 30 31 32 34 def read_exon_positions(self, filename'); if len(filename) - B: filename = self.exon_position_filename else: self.exon_postion_filename = filename self.exon_positions = 1 tuple int(num_str) for num_str in line.split()) for line in open(filename, 'r') 1 def create_mRNA(self): I self.mRNA_seq= for begin, end in self.exon_positions: self. RNA_seq u selt.dna_seq[begin: end).replace('T', 'U') def save_mRNA seq(self, filename", letters_per_line - 70): if len(filename) - 8: filename = self.mRNA_seq_filename else: self.mRNA seq_filename = filename savefile = open(filename, 'W') for 1 in range(0, ten(self. ANA_sea), letters_per_Line): begin = 1 end - begin - letters_per_line savefile.write(self. RNA seqfbegintend) + ' ') savefile.close() def read_translation conversion_map(self, filenanes") it len(filename) - 6: filename - self.translation_conversion_map_filename else: self.translation_conversion_map_filenane = filename self.translation_conversion_map { line.split( 101: line.split(1:) for line in open(filenane, ') } det create protein_seql self): self.protein_seq begin_pos= self. ANA_seq.find('AUG) 54 55 56 57 58 59 60 61 62 64 65 65 67 69 70 begin = 1 end begin + letters_per_line savefile.write(self. ANA_seq[begin:end) + ' ') savefile.close() def read_translation_conversion_map(self, filename=''): if ten(filename) : filename = self.translation_conversion_map_filename es else: self. translation conversion_map_filename-filename selt.translation_conversion_map = { line.split(10: line.split()11:1 for line in open(filename, 'r)) det create_protein_seq(self): self.protein_seq - begin.pos = self. RNA seq.find('AUG) for 1 in range(begin_pos, lentself. RNA sea), 3); triplet self.mRNA seq(1:1+3) aminoacid self.translation_conversion_map(triplet 101 if amino acid w 'X'T break; else: self.protein seq amino acid 82 3 BG 85 86 B7 BB 09 # Homework Week 3, Problem 4 # # Complete this function. 1 + You must not change other parts of this class to keep it work properly. def mutate_dna_seq(self, pos, base): Python 3 C # The following code is provided for your test. + You can modify as you want, to test your function. import Gene import os dna_seq_filename = os.path.join('data', 'lactase_gene.txt') lactase_gene = Gene. Genel lactase') lactase_gene. read_dna_seq_file(dna_seq_filename) print(lactase_gene.dna_seq[100:150]) lactase_gene.mutate_dna_seq(103, 'T) print(lactase_gene.dna_seq(108:150]) CALCCTGGTCCTCTAACCAATGACTTGCTGCACAACCTGAGTGGTCTCC CATCCTGGTCCTCTAACCAATGACTTGCTGCACAACCTGAGTGGTCTCC The following code is provided for your test. + You can nodify it as you want, to test your function. fron Gene import Gene inport os dna_seq_filename - os. path.join('data', 'lactase_gene.txt') lactase_gene - Genel lactose) lactase_gene.read_dna_sectiledna_se_filename) print (lactase_gene.dna seg110011501) lactase gene.mutate dna, seg(103T) print (lactose_gene.dna, seq 100-1501) Python 3 die 10 1 class Gene(): 2 def __init_(self, gene_name, dna_seq_filename=", exon_position_filename=' translation_conversion_map_filename='); mRNA_seq_filename, self.gene_name gene_name selt.dna seg_filename = dna_seq_filename self.exon_position_filename = exon_position_filename self.mRNA seq_filename = mRNA seq_filename self.translation_conversion_map_filename = translation_conversion_map_filename I self.dna_seq=1 21 self.exon_positions - 11 12 self.mRNA_seg - 13 self. translation conversion_map) 14 self.protein_seq=1 15 16 det read_dna_seq_file(self, filename'); 17 it len(filename) - 0: 18 filename = self.dna_seq_filename else: 20 self.dna_seq. Filename - filenane 21 22 lines open(filename, 'r) 23 self.dno_seg - ".joint line.strip() for line in lines) 24 25 def read_mRNA seq.fileself, filename): 26 if len(filename) -- : 27 Filename = self.mRNA_seq_filename 28 else: 29 self. RNA seq_filename = filename Lines = open(filename, '') self. RNA_seq= ".joint line.strip) for line in lines 1) 33 def read_exon_positions(self, filename'): 35 if len(filename) : 36 filename = self.exon_position filename 19 30 31 32 34 def read_exon_positions(self, filename'); if len(filename) - B: filename = self.exon_position_filename else: self.exon_postion_filename = filename self.exon_positions = 1 tuple int(num_str) for num_str in line.split()) for line in open(filename, 'r') 1 def create_mRNA(self): I self.mRNA_seq= for begin, end in self.exon_positions: self. RNA_seq u selt.dna_seq[begin: end).replace('T', 'U') def save_mRNA seq(self, filename", letters_per_line - 70): if len(filename) - 8: filename = self.mRNA_seq_filename else: self.mRNA seq_filename = filename savefile = open(filename, 'W') for 1 in range(0, ten(self. ANA_sea), letters_per_Line): begin = 1 end - begin - letters_per_line savefile.write(self. RNA seqfbegintend) + ' ') savefile.close() def read_translation conversion_map(self, filenanes") it len(filename) - 6: filename - self.translation_conversion_map_filename else: self.translation_conversion_map_filenane = filename self.translation_conversion_map { line.split( 101: line.split(1:) for line in open(filenane, ') } det create protein_seql self): self.protein_seq begin_pos= self. ANA_seq.find('AUG) 54 55 56 57 58 59 60 61 62 64 65 65 67 69 70 begin = 1 end begin + letters_per_line savefile.write(self. ANA_seq[begin:end) + ' ') savefile.close() def read_translation_conversion_map(self, filename=''): if ten(filename) : filename = self.translation_conversion_map_filename es else: self. translation conversion_map_filename-filename selt.translation_conversion_map = { line.split(10: line.split()11:1 for line in open(filename, 'r)) det create_protein_seq(self): self.protein_seq - begin.pos = self. RNA seq.find('AUG) for 1 in range(begin_pos, lentself. RNA sea), 3); triplet self.mRNA seq(1:1+3) aminoacid self.translation_conversion_map(triplet 101 if amino acid w 'X'T break; else: self.protein seq amino acid 82 3 BG 85 86 B7 BB 09 # Homework Week 3, Problem 4 # # Complete this function. 1 + You must not change other parts of this class to keep it work properly. def mutate_dna_seq(self, pos, base): Python 3 C # The following code is provided for your test. + You can modify as you want, to test your function. import Gene import os dna_seq_filename = os.path.join('data', 'lactase_gene.txt') lactase_gene = Gene. Genel lactase') lactase_gene. read_dna_seq_file(dna_seq_filename) print(lactase_gene.dna_seq[100:150]) lactase_gene.mutate_dna_seq(103, 'T) print(lactase_gene.dna_seq(108:150]) CALCCTGGTCCTCTAACCAATGACTTGCTGCACAACCTGAGTGGTCTCC CATCCTGGTCCTCTAACCAATGACTTGCTGCACAACCTGAGTGGTCTCC The following code is provided for your test. + You can nodify it as you want, to test your function. fron Gene import Gene inport os dna_seq_filename - os. path.join('data', 'lactase_gene.txt') lactase_gene - Genel lactose) lactase_gene.read_dna_sectiledna_se_filename) print (lactase_gene.dna seg110011501) lactase gene.mutate dna, seg(103T) print (lactose_gene.dna, seq 100-1501) Python 3 die










