Answered step by step
Verified Expert Solution
Question
1 Approved Answer
in python Problem The GC content of a DNA string is given by the percentage of symbols in the string that are 'C' or 'G'
in python 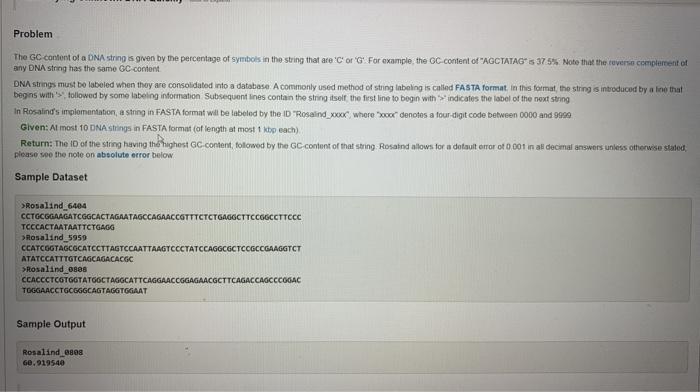
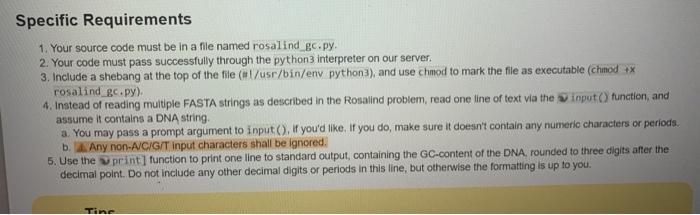
Problem The GC content of a DNA string is given by the percentage of symbols in the string that are 'C' or 'G' For example, the OC content of AGCTATAGS 375% Note that the reverse compartent et any DNA string has the same GC content DNA strings must be labeled when they are consolidated into a database. A commonly used method of string Inbeling is called FASTA format in this format, the string is introduced by a line that begins with followed by some labeling information Subsequentines contain the string itself the first line to begin with indicates the label of the next string In Rosalind's implementation, a string in FASTA format will be labeled by the ID "Rosalind_XXXX Whore" denotes a four-digit code between 0000 and 999 Given: Almost 10 DNA shings in FASTA format (of length at most peach), Return: The 10 of the string having thoughost GC content, followed by the GC content of that string. Rosand allows for a dotauit error of 0.001 in al decimal answers unless otherwise stated, please see the note on absolute error below Sample Dataset Rosalind 6404 CCTGCGGAAGATCGGCACTAGAATAGCCAGAACCOTTTCTCTGAGGCTTCCGOCCTTCCC TCCCACTAATAATTCTGAGG Rosalind_5950 CCATCGGTAGCGCATCCTTAGTCCAATTAAGTCCCTATCCAGGCGCTCCGCCGAAGGTCT ATATCCATTTGTCAGCAGACACGC Rosalind_0828 CCACCCTCOTGGTATGOCTAGGCATTCAGGAACCGGAGAACGCTTCAGACCAGCCCOGAC TOGGAACCTGCGGGCAGTAGGTGGAAT Sample Output Rosalind 6808 60.919540 Specific Requirements 1. Your source code must be in a file named rosalind ge.py. 2. Your code must pass successfully through the python3 interpreter on our server. 3. Include a shebang at the top of the file (l/usr/bin/env python3), and use chmod to mark the file as executable (chmod ** rosalind_gc.py) 4. Instead of reading multiple FASTA strings as described in the Rosalind problem, read one line of text via the input() function, and assume it contains a DNA string. a. You may pass a prompt argument to input(). If you'd like. If you do, make sure it doesn't contain any numeric characters or periods. b. Any non-A/CIG/T input characters shall be ignored. 5. Use the print] function to print one line to standard output, containing the GC-content of the DNA, rounded to three digits after the decimal point. Do not include any other decimal digits or periods in this line, but otherwise the formatting is up to you. Tine Problem The GC content of a DNA string is given by the percentage of symbols in the string that are 'C' or 'G' For example, the OC content of AGCTATAGS 375% Note that the reverse compartent et any DNA string has the same GC content DNA strings must be labeled when they are consolidated into a database. A commonly used method of string Inbeling is called FASTA format in this format, the string is introduced by a line that begins with followed by some labeling information Subsequentines contain the string itself the first line to begin with indicates the label of the next string In Rosalind's implementation, a string in FASTA format will be labeled by the ID "Rosalind_XXXX Whore" denotes a four-digit code between 0000 and 999 Given: Almost 10 DNA shings in FASTA format (of length at most peach), Return: The 10 of the string having thoughost GC content, followed by the GC content of that string. Rosand allows for a dotauit error of 0.001 in al decimal answers unless otherwise stated, please see the note on absolute error below Sample Dataset Rosalind 6404 CCTGCGGAAGATCGGCACTAGAATAGCCAGAACCOTTTCTCTGAGGCTTCCGOCCTTCCC TCCCACTAATAATTCTGAGG Rosalind_5950 CCATCGGTAGCGCATCCTTAGTCCAATTAAGTCCCTATCCAGGCGCTCCGCCGAAGGTCT ATATCCATTTGTCAGCAGACACGC Rosalind_0828 CCACCCTCOTGGTATGOCTAGGCATTCAGGAACCGGAGAACGCTTCAGACCAGCCCOGAC TOGGAACCTGCGGGCAGTAGGTGGAAT Sample Output Rosalind 6808 60.919540 Specific Requirements 1. Your source code must be in a file named rosalind ge.py. 2. Your code must pass successfully through the python3 interpreter on our server. 3. Include a shebang at the top of the file (l/usr/bin/env python3), and use chmod to mark the file as executable (chmod ** rosalind_gc.py) 4. Instead of reading multiple FASTA strings as described in the Rosalind problem, read one line of text via the input() function, and assume it contains a DNA string. a. You may pass a prompt argument to input(). If you'd like. If you do, make sure it doesn't contain any numeric characters or periods. b. Any non-A/CIG/T input characters shall be ignored. 5. Use the print] function to print one line to standard output, containing the GC-content of the DNA, rounded to three digits after the decimal point. Do not include any other decimal digits or periods in this line, but otherwise the formatting is up to you. Tine 

Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


