
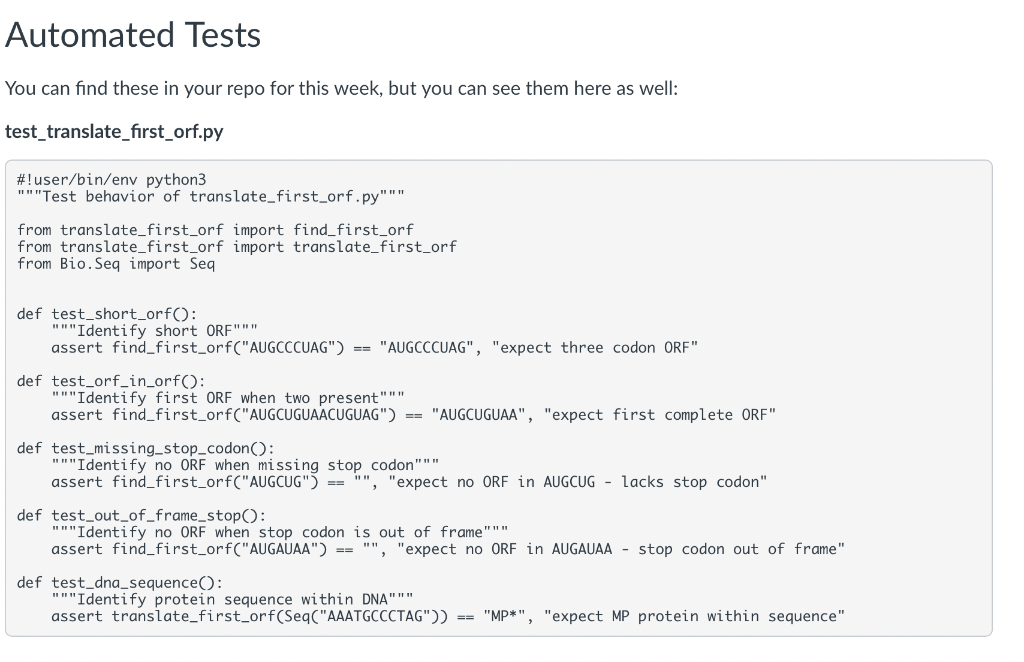
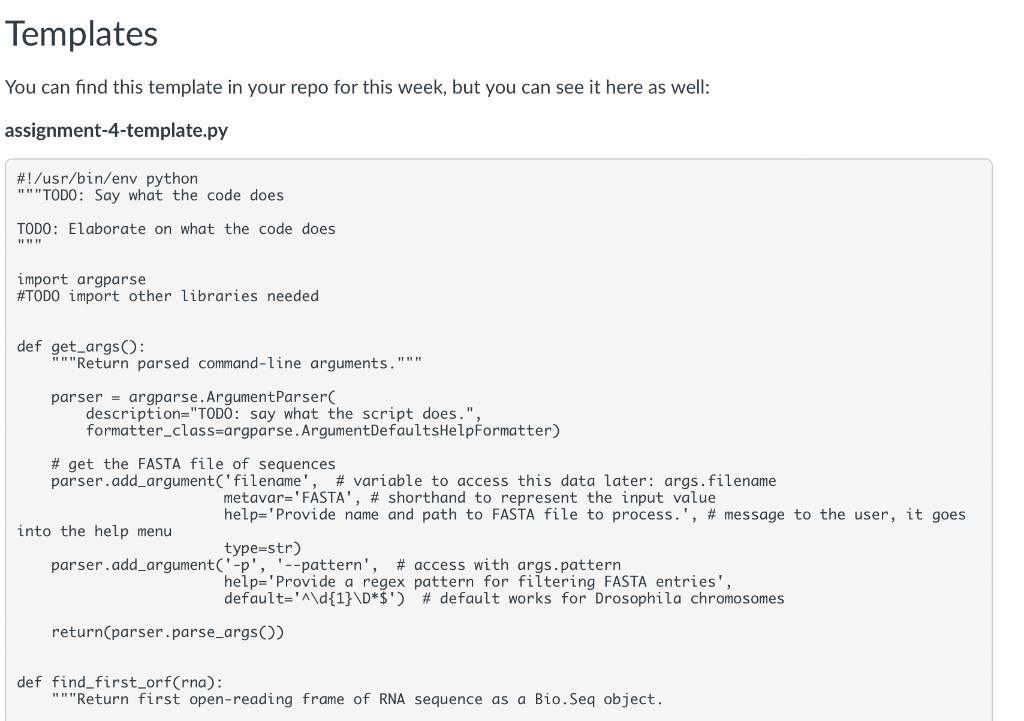
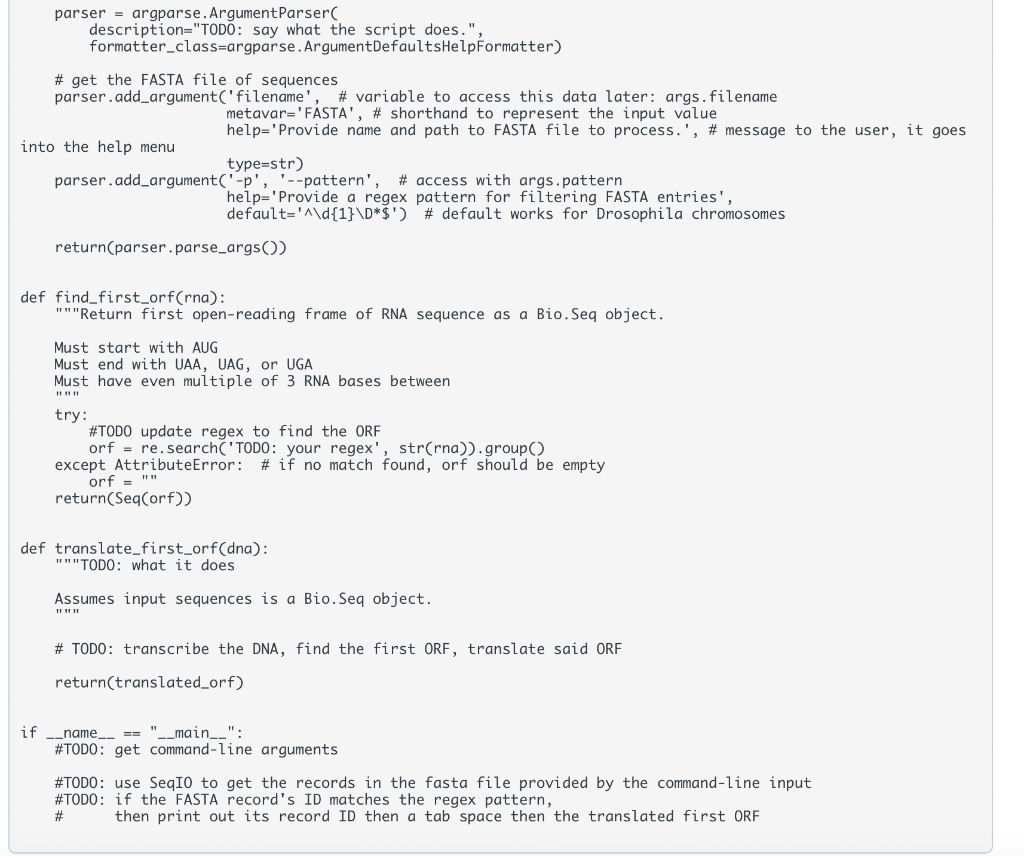
Instructions: - Work through Object-Oriented Programming \& Biopython and Practice Finding Open Reading Frames to gain the building blocks to complete this assignment - Review the spec below for the script we would like you to create - Review the automated tests in the folder to understand what we will be testing. Be sure you know the intent of the tests; when grading your assignments. We will also use a secondary set of tests that you haven't see but that test the same behaviors. - Write your functions, comments, and any supporting documentation requested in the spec. (You may use the provided template to help or write from scratch; in both cases, it is up to you to ensure they pass the tests.) Functions: : takes an RNA sequence and returns the first complete ORF as a Seq object - ORFs are defined here as in the practice sections by having an "AUG" start codon, some iteration of codons containing A, C, U, and G, ending in one of the following stop codons (UAA, UAG, or UGA), and having no internal stop codons : takes a DNA sequence, transcribes it into RNA, finds the first ORF, translates said ORF into a protein, and returns that protein - Use this area to handle user input, open FASTA files, decide which FASTA entries to process, and print the results to the command line. - Output should follow the format of: The functions should work just as well with any genome, not just the Drosophila one. Manual Tests Make sure you can provide the Drosophila genome FASTA file ("/work/courses/BINF6308/data_BINF6308/Module4/dmel-all-chromosome-r6.17.fasta") and get the following result: 2L:2R:3L:3R:4:MHDRGSRTDI*MSF*MIAYARVVPTYCAL*MGPSEPSTEPSTGPVRDQYGTSTGPVRDQYGTSTGPVRDQYGTSTGPVRDQYGTSTGPSTEPSTGPVRDQYGTSTGPVRD*MNGIIIGNSTIFNNLYQSTNLVNALLIYLT* Automated Tests You can find these in your repo for this week, but you can see them here as well: test_translate_first_orf.py \#!user/bin/env python 3 "" "Test behavior of translate_first_orf.py"" from translate_first_orf import find_first_orf from translate_first_orf import translate_first_orf from Bio.Seq import Seq def test_short_orf(): "" "Identify short ORF""" assert find_first_orf("AUGCCCUAG") == "AUGCCCUAG", "expect three codon ORF" def test_orf_in_orf(): "" Identify first ORF when two present"" assert find_first_orf("AUGCUGUAACUGUAG") == "AUGCUGUAA", "expect first complete ORF" def test_missing_stop_codon(): "" "Identify no ORF when missing stop codon""" assert find_first_orf("AUGCUG") == "", "expect no ORF in AUGCUG - lacks stop codon" def test_out_of_frame_stop() : "" "Identify no ORF when stop codon is out of frame"" " assert find_first_orf("AUGAUAA") == "", "expect no ORF in AUGAUAA - stop codon out of frame" def test_dna_sequence(): "" Identify protein sequence within DNA""" assert translate_first_orf(Seq("AAATGCCCTAG")) == "MP*", "expect MP protein within sequence" You can find this template in your repo for this week, but you can see it here as well: assignment-4-template.py \#!/usr/bin/env python "" TOD0: Say what the code does TOD0: Elaborate on what the code does import argparse \#TODO import other libraries needed def get_args(): ""Return parsed command-line arguments." " parser = argparse. ArgumentParser C description="TODO: say what the script does.", formatter_class=argparse.ArgumentDefaultsHelpFormatter) \# get the FASTA file of sequences parser.add_argument('filename', \# variable to access this data later: args.filename metavar='FASTA', \# shorthand to represent the input value help='Provide name and path to FASTA file to process.', \# message to the user, it goes into the help menu type =str) parser.add_argument('- p, ,'-pattern', \# access with args.pattern help='Provide a regex pattern for filtering FASTA entries', default t=d{1}\D$) \# default works for Drosophila chromosomes return(parser.parse_args()) def find_first_orf(rna): "" Return first open-reading frame of RNA sequence as a Bio.Seq object











