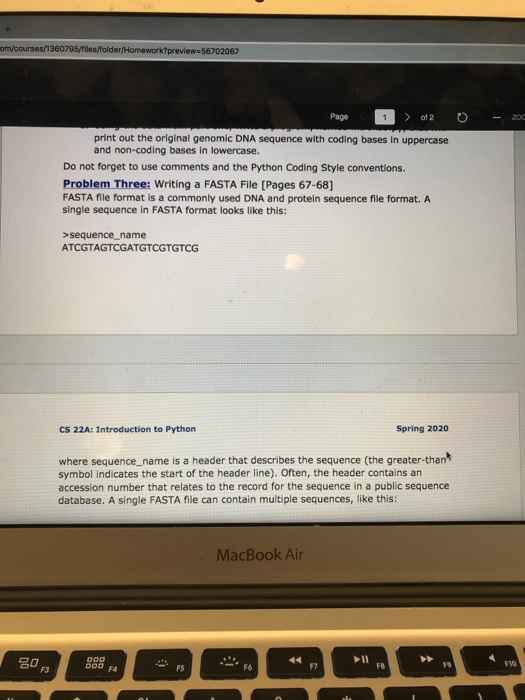
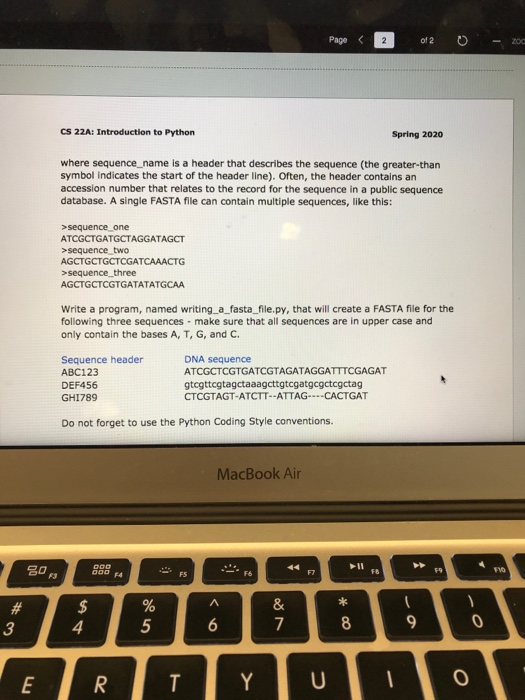
om/courses/1360795/files/folder/Homework?preview=56702067 - 200 Page 1 > of 2 0 print out the original genomic DNA sequence with coding bases in uppercase and non-coding bases in lowercase. Do not forget to use comments and the Python Coding Style conventions. Problem Three: Writing a FASTA File [Pages 67-68] FASTA file format is a commonly used DNA and protein sequence file format. A single sequence in FASTA format looks like this: >sequence_name ATCGTAGTCGATGTCGTGTCG CS 22A: Introduction to Python Spring 2020 where sequence_name is a header that describes the sequence (the greater than symbol indicates the start of the header line). Often, the header contains an accession number that relates to the record for the sequence in a public sequence database. A single FASTA file can contain multiple sequences, like this: MacBook Air Page sequence_one ATCGCTGATGCTAGGATAGCT >sequence_two AGCTGCTGCTCGATCAAACTG >sequence_three AGCTGCTCGTGATATATGCAA Write a program, named writing_a_fasta_file.py, that will create a FASTA file for the following three sequences - make sure that all sequences are in upper case and only contain the bases A, T, G, and C. Sequence header ABC123 DEF456 GHI789 DNA sequence ATCGCTCGTGATCGTAGATAGGATTTCGAGAT gtcgttogtagctaaagcttgtcgatgcgctcgctag CTCGTAGT-ATCTT--ATTAG----CACTGAT Do not forget to use the Python Coding Style conventions. MacBook Air om/courses/1360795/files/folder/Homework?preview=56702067 - 200 Page 1 > of 2 0 print out the original genomic DNA sequence with coding bases in uppercase and non-coding bases in lowercase. Do not forget to use comments and the Python Coding Style conventions. Problem Three: Writing a FASTA File [Pages 67-68] FASTA file format is a commonly used DNA and protein sequence file format. A single sequence in FASTA format looks like this: >sequence_name ATCGTAGTCGATGTCGTGTCG CS 22A: Introduction to Python Spring 2020 where sequence_name is a header that describes the sequence (the greater than symbol indicates the start of the header line). Often, the header contains an accession number that relates to the record for the sequence in a public sequence database. A single FASTA file can contain multiple sequences, like this: MacBook Air Page sequence_one ATCGCTGATGCTAGGATAGCT >sequence_two AGCTGCTGCTCGATCAAACTG >sequence_three AGCTGCTCGTGATATATGCAA Write a program, named writing_a_fasta_file.py, that will create a FASTA file for the following three sequences - make sure that all sequences are in upper case and only contain the bases A, T, G, and C. Sequence header ABC123 DEF456 GHI789 DNA sequence ATCGCTCGTGATCGTAGATAGGATTTCGAGAT gtcgttogtagctaaagcttgtcgatgcgctcgctag CTCGTAGT-ATCTT--ATTAG----CACTGAT Do not forget to use the Python Coding Style conventions. MacBook Air