Question
Please help! I am trying to find a potential gene by start and stop codons which I have already found. The start must start with
Please help! I am trying to find a potential gene by start and stop codons which I have already found. The start must start with ATG and can end with TAG, TAA, TGA. Now I just need to somehow combine the start codon, substring, and stop codons all in one string. My code so far.
int main()
{
string fileName;
cout
cin >> fileName;
fstream fileStream(fileName, ios::in);
if (fileStream.fail())
{
cout
exit(1);
}
cout
char dummyChar = '>';
fileStream >> dummyChar;
string dummyString;
getline(fileStream, dummyString);
cout
cout
string genome,fasta;
int length;
float cTotal, gTotal, gcTotal, gcPercent;
while (!fileStream.eof())
{
getline(fileStream,genome); // Saves the line in genome.
fasta += genome;//appending each line to string fasta
}
length = fasta.length();
cTotal = countGenes(fasta, 'C'); // Acquires total amount of "C" genes
gTotal = countGenes(fasta, 'G'); // Acquires total amount of "G" genes
gcTotal = (gTotal + cTotal); // Total amount of both "C" and "G" genes
gcPercent = ((gcTotal / fasta.size()) * 100); // Total percentage of both "C" and "G" genes out of all genes
cout
vector startCodonPositions;
startCodon(fasta, startCodonPositions, "ATG");
cout
vector stopCodonPositions;
startCodon(fasta, stopCodonPositions, "TAG");
startCodon(fasta, stopCodonPositions, "TGA");
startCodon(fasta, stopCodonPositions, "TAA");
cout
return 0;
}
My gene.cpp
int countGenes(string str, char gene)
{
int count = 0, length = 7374; // Counter and string length
for (unsigned i = 0; i
{
if (str[i] == gene)
count++;
}
// See how many times the character repeats
int geneRepetition = length / str.size();
count = (count * geneRepetition);
/* If the length is not a multiple of the string size
check for the remaining repeating characters. */
for (unsigned i = 0; i
{
if (str[i] == gene)
count++;
}
return count;
}
void startCodon(string str, vector& vec, string codon)
{
size_t start = 0;
bool search = true;
while (search)
{
start = str.find(codon, start);
if (start == string::npos)
break;
vec.push_back(start);
start++;
}
}
Output:
How to find a potential gene
Part of the csv file:
>NC_003988.1 Simian enterovirus A, complete genome GAGTGTTCCCACCCAACAGGCCCACTGGGTGTTGTACTCTGGTATTACGGTACCTTTGTACGCCTATTTT ATTTCCCCCCCCTTTTTGAAACTTAGAAGTTAATAATAAACACGCTCACTAGGTGCACTACATCCAGTAG TGTAATGAGCAAGCACTTCTGTCTYCCCCGGGAGGGATATATGGTACGCTGTGCAAACGGCGGAAATTAA TCCTACCGTTAACCGCCCACCTACTCCGAGAAGCCTAGTACCTAATTGGATTTATCAATGGAGTTGCGCT CAGCAGGTGACCCTGACCTGCCAGCTCCGGCTGATGGACCTGGGCTTTCCCCACAGGCGACTGTGGCCCA GGTCGCGTGGCGGCCGGCCCACCCCCCTGGGTGGGACGCCTTGATAATGACAAGGTGGGAAGAGCCTATT GGGCTAGCTGGTTTCCTCCGGCCTCCTGAATGCGGCTAACCTTAACCCCAGAGCATATGGTAGCAACCCA GCTACTAGTATGTCATAATGCGTAAGTCTGGGATGGGACCGACTACTTTGGAGAGTCCGTGTTTCTATTG TTTCTTTAATCAATCTTATGGTGACAATTTATAGTGCCCTGAGTATTGATTGGTTGTTGCTTTTGACAAT TATTGAGACATCACATAGACATAATGGGAGCTCAAGTAAGCAGGCAAACGTCTGGTGCGCATGACACCCG GATACGGGCTGAACAGGGCGCAAACATTCATTATACTAATATCAATTATTATAGAGATGCAGCTAGCAAT GCAGCAAGCAAAATGGACTATTCCCAGGATCCGGACAAGTTCACGAAACCAGTACTTGATGCTATAACTG AACCATTACCCACGCTGAAGTCCCCTAGTGCTGAGGCATGTGGGTACAGCGACCGAGTTGCACAACTGAC AATTGGCAATTCCACTATCACTACTCAGGAAGCCGCCAATGTGGTGGTCGCATATGGACAATGGCCTGAA TATTTAGATTCGAAGGATGCAACTGCCGTGGATAAGCCCACACAGCCCGATGTAGCCTCAAATAGATTTT ACACTCTTAAGACAGTGTCTTGGGAGAAGAGTTCAACTGGCTGGTATTGGAAATTCTCGGATTGTCTGGC TTCTGTTGGATTATTTGGACAGAATGTACAGTATCATTATTTAGGCCGTTATGGGTTAGCGGTTCATGTG CAATGTAATGCTTCAAAATTTCATCAGGGCACTCTACTGGTCTTAGCAATACCAGAATGGGAGATTGGGG TGTCTAATGCTGATAGGGCATCCTTTAATCTAACAAACCCCGATAAGAACGGGCATACTATGACTGGTCA AGAAGCTTATTGCTTACATAATGGGACTAACATCCATTCTTCACTGGTATTTCCACATCAATTCATCAAT CTTAGGACAAACAATTGTGCTACGTTAGTCTTGCCCTATGTGGGAGCAACACCACTGGACACACCGATCA AGCATAATGTTTGGTCATTGGTAGTAATACCGGTGGTCCCGTTGGATTACACCACTGGTGCAACTACACA AGTGCCTATAACAATAACAATGGCTCCAATGGCGTGCGAGTTTAACGGACTGCGCAATGCCATCACCCAA GGGCTGCCAGTACTCAATACACCCGGCTCTGGGCAGTTTGTGACTACAGATAATTTCCAATCACCAAACT TGATTCCAAATTTTGATGTGACACAAGTCTTTAATAGTCCAGGTGAAATTATTAATTTACAGCAGTATGT CCAGATTGAGGGCATTATGGAAATCAATAATGTAGCAAGTGCAAATAATTTGGAGAGAATTCGCATTCCA ATATCAGTCCAGAGTGGAATTGATGAGATGTTATTTGCAATCAACTGCAACCCAGGAACAGCCCAGGAGT TTAGACGCACACCCCTGGGAGATGTGTGTAGGTATTATACACAGTGGTCAGGTAGCATACAAATTACATT TACATTTTGTGGTTCATTTATGACAACAGGAAAATTATTAATTTGCTACACCCCTCCGGGTGGTCGAGTA CCACAAAATAGAGAGGAGGCAATGCTAGGGACTAATGTGATCTGGGATTTTGGTTTACAATCCAGCGTTA CGCTGAACATACCGTGGATAAGTGGAGCCCATTTTAGAAACACTTCTGTTAATGTCGATGGTTTTGATAA CACAGGGTATGTATCTGCTTGGTTTCAAACGAACATGGTAGTTCCTCCCGATGCTCCAACGACTGCTTAT ATATTGGCTTTTACATCAGCCAAGGATGATTTCTCGATGCGCTTGTTGCGGGATACAGCAGAGATTTCGC AAGACGGATTTCTGCAAGGACCAATAGATCAAGCAATAGAAAAAGTAATCACTGATGTAGTGTCTGACAC GCGTGAGTCTAGTAGTGACTTTAGCATTGGGGCTGTTCCAGCATTGAATGCGGTGGAAACTGGAGCCACT TCGCAAGCTAGTGTTGAGTCCACCATTGAGACGCGGGCCGTGCAGAATCGTCATCGCACTTCTGAGATGA GCGTGGAAAGCTTTTTGGGCCGCTCTAGTTTAGTAACTCGCTTTACCATTAATAATGGAGGAACAAATAA TGCCACGAAGTTTCGTAACTGGAAAATAAACTTAAAGGAAGTGGTGCAGCTGCGGCGTAAATTAGAAATG TTTACTTACGTGCGCTTTGATCTTGAGGTGACTATAGTGGCTGTGAATTTGACTGGAAATGGAGGAGTGC GTTACATGTACCAAGCAATGTACTGCCCCCCAGGTGCCCCCCTCCCCACCAATGCTGATCAATATCTGTG GCAATCCTCGACAAATCCCTCCATAATCGGAGCAGTTGGTGAAGTCCCAGGCAGAGTATCAGTGCCTTTT GTGTCAAATGCTAATATGTATGCCACCTTTTATGATGGATATCCATCCTTTGGAAGCATAAATGGACAGG GAAATGGCTCTGATTACGGTGCATTCATACCAAATGATATGGGTACATTGTGTTTCCGATTACTCAATAT CTTTAATAATGGTCCACAAATTCAATTTAGAGTGTTCATGAAACCCAAGCATGTACGAGTATGGTGCCCA
Thank you.
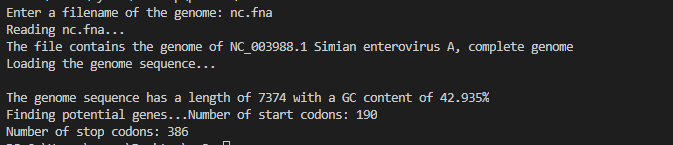
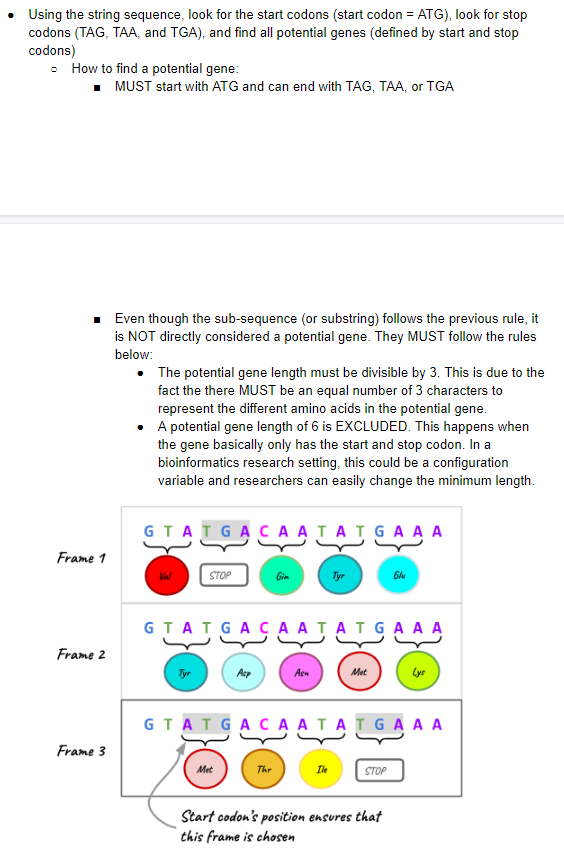
Enter a filename of the genome: nc.fna Reading nc.fna... The file contains the genome of NC_003988.1 Simian enterovirus A, complete genome Loading the genome sequence... The genome sequence has a length of 7374 with a GC content of 42.935% Finding potential genes...Number of start codons: 190 Number of stop codons: 386 Using the string sequence, look for the start codons (start codon = ATG), look for stop codons (TAG, TAA, and TGA), and find all potential genes (defined by start and stop codons) How to find a potential gene: MUST start with ATG and can end with TAG, TAA, or TGA Even though the sub-sequence (or substring) follows the previous rule, it is NOT directly considered a potential gene. They MUST follow the rules below: The potential gene length must be divisible by 3. This is due to the fact the there MUST be an equal number of 3 characters to represent the different amino acids in the potential gene. A potential gene length of 6 is EXCLUDED. This happens when the gene basically only has the start and stop codon. In a bioinformatics research setting, this could be a configuration variable and researchers can easily change the minimum length. GTA IGA CA A LA GA A A Frame 1 STOP Gin Frame 2 GTA GACAAJ ATG A A A Pop TH Mie Guy GTATGACA ATALGA A A Frame 3 Met | STOP Start codon's position ensures that this frame is chosen Enter a filename of the genome: nc.fna Reading nc.fna... The file contains the genome of NC_003988.1 Simian enterovirus A, complete genome Loading the genome sequence... The genome sequence has a length of 7374 with a GC content of 42.935% Finding potential genes...Number of start codons: 190 Number of stop codons: 386 Using the string sequence, look for the start codons (start codon = ATG), look for stop codons (TAG, TAA, and TGA), and find all potential genes (defined by start and stop codons) How to find a potential gene: MUST start with ATG and can end with TAG, TAA, or TGA Even though the sub-sequence (or substring) follows the previous rule, it is NOT directly considered a potential gene. They MUST follow the rules below: The potential gene length must be divisible by 3. This is due to the fact the there MUST be an equal number of 3 characters to represent the different amino acids in the potential gene. A potential gene length of 6 is EXCLUDED. This happens when the gene basically only has the start and stop codon. In a bioinformatics research setting, this could be a configuration variable and researchers can easily change the minimum length. GTA IGA CA A LA GA A A Frame 1 STOP Gin Frame 2 GTA GACAAJ ATG A A A Pop TH Mie Guy GTATGACA ATALGA A A Frame 3 Met | STOP Start codon's position ensures that this frame is chosenStep by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


