
PLEASE HELP THIS MAKES NO SENSE
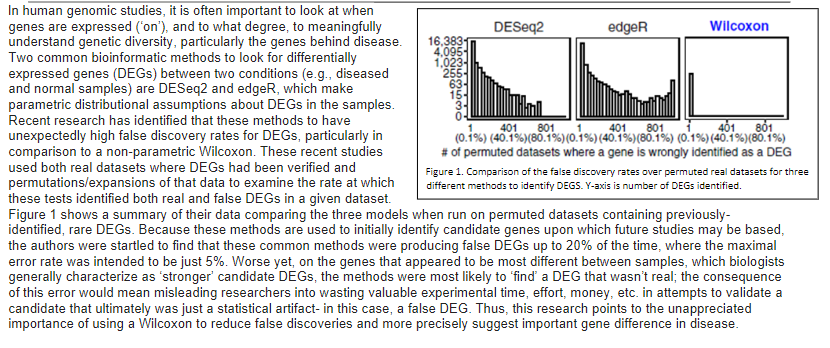

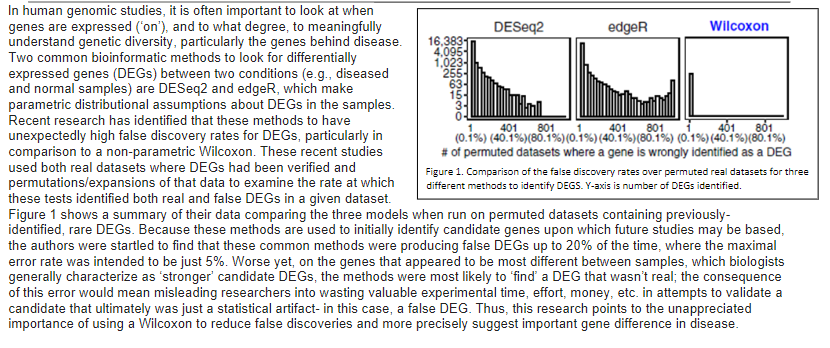
4,095- 255- 63 15- In human genomic studies, it is often important to look at when genes are expressed ('on'), and to what degree, to meaningfully DESeq2 edger Wilcoxon understand genetic diversity, particularly the genes behind disease. 16,383 Two common bioinformatic methods to look for differentially 1,023- expressed genes (DEGs) between two conditions (e.g., diseased and normal samples) are DESeq2 and edge, which make parametric distributional assumptions about DEGs in the samples. Recent research has identified that these methods to have 401 801 unexpectedly high false discovery rates for DEGs, particularly in (0.1%) (40.4%y(80.1%)(0.1%) (40.1%)(80.1%) (0.1%)(40.1%)(80.1%) comparison to a non-parametric Wilcoxon. These recent studies # of permuted datasets where a gene is wrongly identified as a DEG used both real datasets where DEGs had been verified and Figure 1. Comparison of the false discovery rates over permuted real datasets for three permutations/expansions of that data to examine the rate at which different methods to identify DEGS. Y-axis is number of DEGs identified. these tests identified both real and false DEGs in a given dataset. Figure 1 shows a summary of their data comparing the three models when run on permuted datasets containing previously- identified, rare DEGs. Because these methods are used to initially identify candidate genes upon which future studies may be based, the authors were startled to find that these common methods were producing false DEGs up to 20% of the time, where the maximal error rate was intended to be just 5%. Worse yet, on the genes that appeared to be most different between samples, which biologists generally characterize as 'stronger' candidate DEGs, the methods were most likely to find a DEG that wasn't real; the consequence of this error would mean misleading researchers into wasting valuable experimental time, effort, money, etc. in attempts to validate a candidate that ultimately was just a statistical artifact- in this case, a false DEG. Thus, this research points to the unappreciated importance of using a Wilcoxon to reduce false discoveries and more precisely suggest important gene difference in disease. 3. Today, it is easier to sample lots of individuals as technology has vastly improved. Further, larger sample sizes upon which to draw makes it easier to create permuted datasets to check each method, without needing to rely on the data meeting each model's assumptions. a. One way the authors checked the models was by intentionally creating datasets that 1) *should* and 2) should not have a DEG identified from comparisons of diseased and normal samples. What are these two datasets each functioning as in their research? Number your responses. b. Using an alpha level of 0.05, state the statistical outcome of a Wilcoxon test of this data for the 2 groups in part A of this question. Number and state in each response what the p value would need to be the outcome of the test with respect to the null hypothesis, and what one could statistically conclude from the test. 4,095- 255- 63 15- In human genomic studies, it is often important to look at when genes are expressed ('on'), and to what degree, to meaningfully DESeq2 edger Wilcoxon understand genetic diversity, particularly the genes behind disease. 16,383 Two common bioinformatic methods to look for differentially 1,023- expressed genes (DEGs) between two conditions (e.g., diseased and normal samples) are DESeq2 and edge, which make parametric distributional assumptions about DEGs in the samples. Recent research has identified that these methods to have 401 801 unexpectedly high false discovery rates for DEGs, particularly in (0.1%) (40.4%y(80.1%)(0.1%) (40.1%)(80.1%) (0.1%)(40.1%)(80.1%) comparison to a non-parametric Wilcoxon. These recent studies # of permuted datasets where a gene is wrongly identified as a DEG used both real datasets where DEGs had been verified and Figure 1. Comparison of the false discovery rates over permuted real datasets for three permutations/expansions of that data to examine the rate at which different methods to identify DEGS. Y-axis is number of DEGs identified. these tests identified both real and false DEGs in a given dataset. Figure 1 shows a summary of their data comparing the three models when run on permuted datasets containing previously- identified, rare DEGs. Because these methods are used to initially identify candidate genes upon which future studies may be based, the authors were startled to find that these common methods were producing false DEGs up to 20% of the time, where the maximal error rate was intended to be just 5%. Worse yet, on the genes that appeared to be most different between samples, which biologists generally characterize as 'stronger' candidate DEGs, the methods were most likely to find a DEG that wasn't real; the consequence of this error would mean misleading researchers into wasting valuable experimental time, effort, money, etc. in attempts to validate a candidate that ultimately was just a statistical artifact- in this case, a false DEG. Thus, this research points to the unappreciated importance of using a Wilcoxon to reduce false discoveries and more precisely suggest important gene difference in disease. 3. Today, it is easier to sample lots of individuals as technology has vastly improved. Further, larger sample sizes upon which to draw makes it easier to create permuted datasets to check each method, without needing to rely on the data meeting each model's assumptions. a. One way the authors checked the models was by intentionally creating datasets that 1) *should* and 2) should not have a DEG identified from comparisons of diseased and normal samples. What are these two datasets each functioning as in their research? Number your responses. b. Using an alpha level of 0.05, state the statistical outcome of a Wilcoxon test of this data for the 2 groups in part A of this question. Number and state in each response what the p value would need to be the outcome of the test with respect to the null hypothesis, and what one could statistically conclude from the test










