Question
REPLACE ALL #TODO WITH THE REQUIRED FUNCTIONS AND ADD COMMENTS THAT ARE REQUIRED Name of the script: interleaved.py Functions: get_args : gets command-line arguments for
REPLACE ALL #TODO WITH THE REQUIRED FUNCTIONS AND ADD COMMENTS THAT ARE REQUIRED
Name of the script: interleaved.py
Functions:
get_args: gets command-line arguments for input and output files using the argparse library
Input: None to the function
Output: the argument object created by the parser.parse_args() call
Additional notes: you will want to get the following from the user via command-line input (hint: each of these should be a parser.add_argument() call)
Path and filename for the left or R1 FASTQ mate-pair file (e.g., Aip02.R1.fastq); should be required!
Path and filename for the right or R2 FASTQ mate-pair file (e.g., Aip02.R2.fastq); should be required!
Path and filename for the output FASTA file name (e.g., Aip02.interleaved.fasta)
Path to the folder for storing logs (default is results/logs/ assuming you run the script from your repo folder)
Base name for the log file (default is the name of the script)
You can leverage the get_args() boilerplate in the template below to get the framework for what you need. This boilerplate includes definitions for the output file and the two logging-related command-line arguments.
interleave: interleaves mate-pair FASTQ sequences into a single FASTA file
Input: SeqIO.parse() iterators for the R1 (left) and R2 (right) FASTQ files (hint: this means you call SeqIO.parse()outside of the function and pass the result of SeqIO.parse() to the function.)
Output: a list (aka array) of SeqRecord objects in interleaved format with first the R1 record for a read, then its R2 record mate, then the next R1 read, then its R2 mate, etc.
Assumptions: Read 1 on the R1 file is the mate of Read 1 in the R2 file.
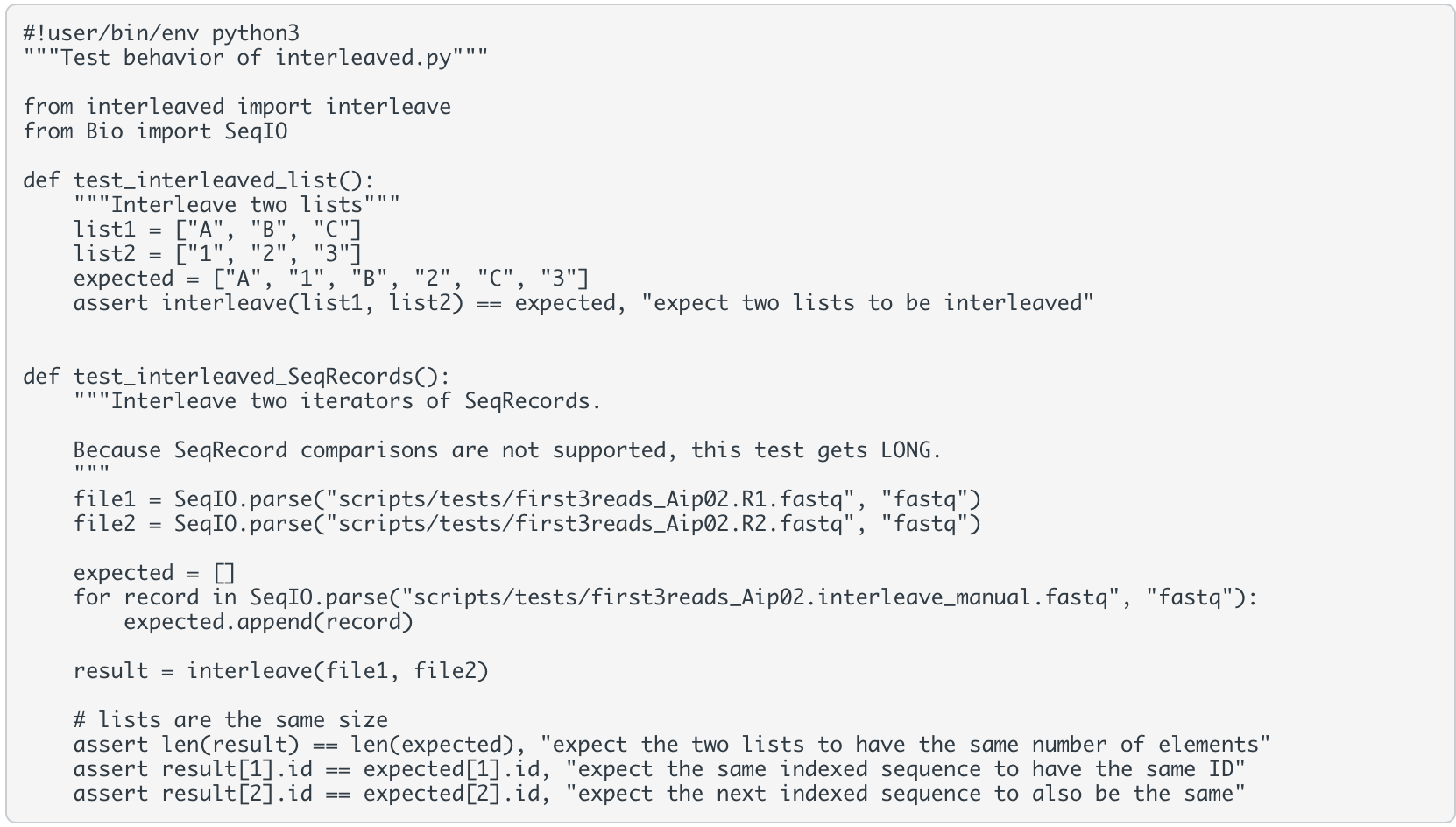
you will want to ensure that the interleaved.py script you write passes both of these tests. test_interleaved.py
As you copy the template, be sure to rename it to interleaved.py and for each "TODO" tag, replace the tag with the request following "TODO." For instance, replace the """TODO: Say what the script does""" DocString with a DocString that says what the script does.
SCRIPT:
#!/usr/bin/env python3 """TODO: Say what the script does""" import argparse # for command-line argument parsing from datetime import datetime # for getting current timestamp from Bio import SeqIO # for reading/writing FASTQ/A files def get_args(): """Return parsed command-line arguments.""" parser = argparse.ArgumentParser( description="Interleave mate-pair FASTQ sequences into a single FASTA file.", formatter_class=argparse.ArgumentDefaultsHelpFormatter) # TODO add argument to get the first mate FASTQ file name (or path) # TODO add argument to get the second mate FASTQ file name # Get output FASTA file name parser.add_argument('-o', '--output', # variable to access this data later: args.output metavar='FASTA', # shorthand to represent the input value help='Provide the path for the output FASTA file.', # message to the user, it goes into the help menu type=str, required=True) # extra arguments to help us format our log file output parser.add_argument('--logFolder', # variable to access this data later: args.logFolder help='Provide the folder for log files.', # message to the user, it goes into the help menu type=str, default="results/logs/") parser.add_argument('--logBase', # variable to access this data later: args.logBase help='Provide the base for the log file name', type=str, default=parser.prog) # get the name of the script return(parser.parse_args()) def pathLogFile(logFolder, logBase): """Return a log file path and name using the current time and script name.""" timestamp = datetime.now().strftime("%Y-%m-%d-%H%M") # get current time in YYYY-MM-DD-HHMM return(f"{logFolder}{timestamp}_{logBase}.log") def interleave(mate1, mate2): """Return list of interleaved SeqRecords. Assumes mate1 and mate2 inputs are SeqIO.parse iterator objects. """ interleaved = [] # TODO: populate the interleaved list with interleaved SeqRecord objects return(interleaved) def logInterleave(args): """Create log of Interleave progress.""" logFile = pathLogFile(args.logFolder, args.logBase) with open(logFile, 'w') as log: log.write(f"Running interleaved.py on {datetime.now()} ") log.write(" **** Summary of arguments ****") # TODO log the two mate files and the output file log.write(" ") # add some space between argument data and the rest of the log # TODO add log lines and commands to do the following steps. # Unsure what/how to log? # I've provided a sample of my log file in the results/logs/2022-10-13-1544_interleaved.py.log file in this repo # 1. Get the FASTQ sequences with SeqIO.parse # 2. Get the interleaved list of SeqRecord objects # 3. Write the interleaved list of SeqRecord objects to our FASTA file with SeqIO.write log.write(f" Script has finished at {datetime.now()}") if __name__ == "__main__": logInterleave(get_args()) # pass arguments directly into the primary function \#!user/bin/env python 3 "" "Test behavior of interleaved.py"" from interleaved import interleave from Bio import SeqI0 def test_interleaved_list(): "" "Interleave two lists"" list1 = ["A", "B", "C"] list2 = ["1", "2", "3"] expected = ["A", "1", "B", "2", "C", "3"] assert interleave(list1, 'ist2) == expected, "expect two lists to be interleaved" def test_interleaved_SeqRecords(): "" Interleave two iterators of SeqRecords. Because SeqRecord comparisons are not supported, this test gets LONG. file1 = SeqI0.parse("scripts/tests/first3reads_Aip02.R1.fastq", "fastq") file2 = SeqI0.parse("scripts/tests/first3reads_Aip02.R2.fastq", "fastq") expected =[] for record in SeqI0.parse("scripts/tests/first3reads_Aip02. interleave_manual. fastq", "fastq"): expected. append(record) result = interleave(file1, file2) \# lists are the same size assert len(result) == len(expected), "expect the two lists to have the same number of elements" assert result[1].id == expected [1].id, "expect the same indexed sequence to have the same ID" assert result[2].id == expected[2].id, "expect the next indexed sequence to also be the same
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


