Question: The figure below shows the recognition sequences and cleavage positions of three restriction enzymes. BamHI Bg/II NdeIl 5'- GGATCC - 3' 3'- CCTAGG -
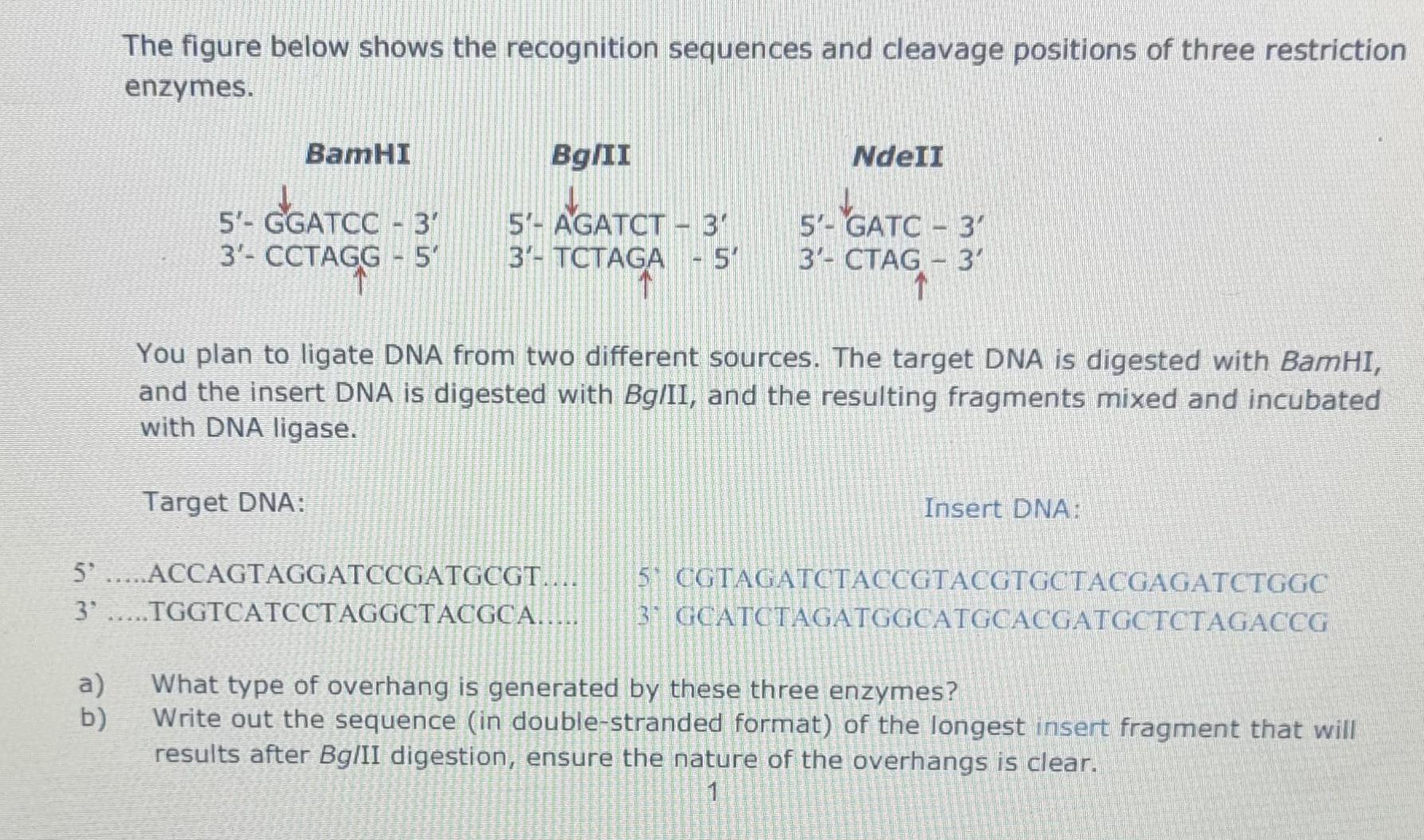

The figure below shows the recognition sequences and cleavage positions of three restriction enzymes. BamHI Bg/II NdeIl 5'- GGATCC - 3' 3'- CCTAGG - 5' 5'- AGATCT - 3" 3'- GA -5' 5- GATC - 3 3- CTAG - 3' You plan to ligate DNA from two different sources. The target DNA is digested with BamHI, and the insert DNA is digested with Bg/II, and the resulting fragments mixed and incubated with DNA ligase. Target DNA: Insert DNA: 5 .....ACCAGTAGGATCCGATGCGT... 5 CGTAGATCTACCGTACGTGCTACGAGATCTGGC 3' .TGGTCATCCTAGGCTACGCA... 31 GCATCTAGATGGCATGCACGATGCTCTAGACCG a) What type of overhang is generated by these three enzymes? b) Write out the sequence (in double-stranded format) of the longest insert fragment that will results after BglII digestion, ensure the nature of the overhangs is clear. 1 c) What is the function of the DNA ligase enzyme? Write out the sequence (in double-stranded format) of the ligation product, with the insert fragment joined into the BamHI site of the target DNA. Use black for target sequences, and blue for insert sequences. Assume the ligation reaction was successful and you have generated a recombinant DNA e) molecule. Which of the three enzymes listed above can be used to excise the insert DNA from the target? Motivate your answer.
Step by Step Solution
3.39 Rating (165 Votes )
There are 3 Steps involved in it
3 a The type of overhang generated from the above restriction enzymes will be of the staggered type ... View full answer

Get step-by-step solutions from verified subject matter experts


