Answered step by step
Verified Expert Solution
Question
1 Approved Answer
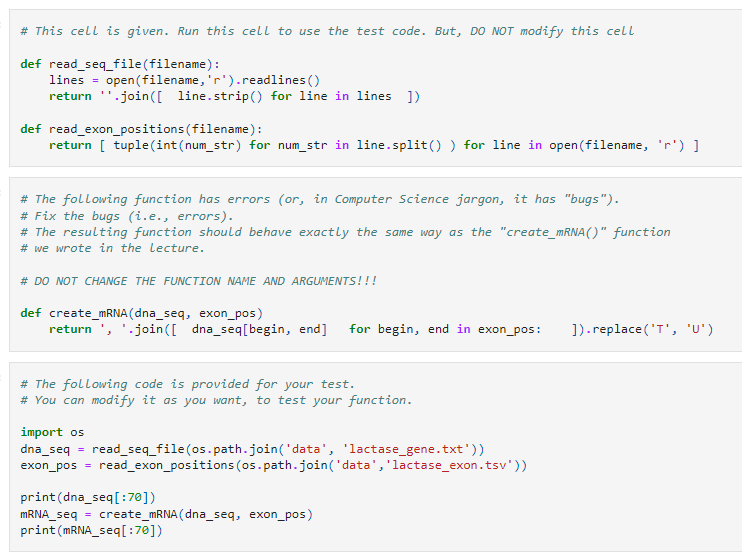
The following corresponds to python programming. Will upvote, thx. 1) We are simplifying the create_mRNA() function. Specifically, we want to implement it in a single
The following corresponds to python programming. Will upvote, thx.
1) We are simplifying the create_mRNA() function. Specifically, we want to implement it in a single line. But there are errors. Find what went wrong and fix them.
a) PLEASE WRITE THE COMPLETED CODE BELOW IN YOUR ANSWER.
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


