The GFF3 format is a commonly used one in bioinformatics for representing sequence annotation. You can find the specification here:
http://www.sequenceontology.org/gff3.shtml
I've placed the genome and annotation for Saccharomyces cerevisiae S288C, on the class server here:
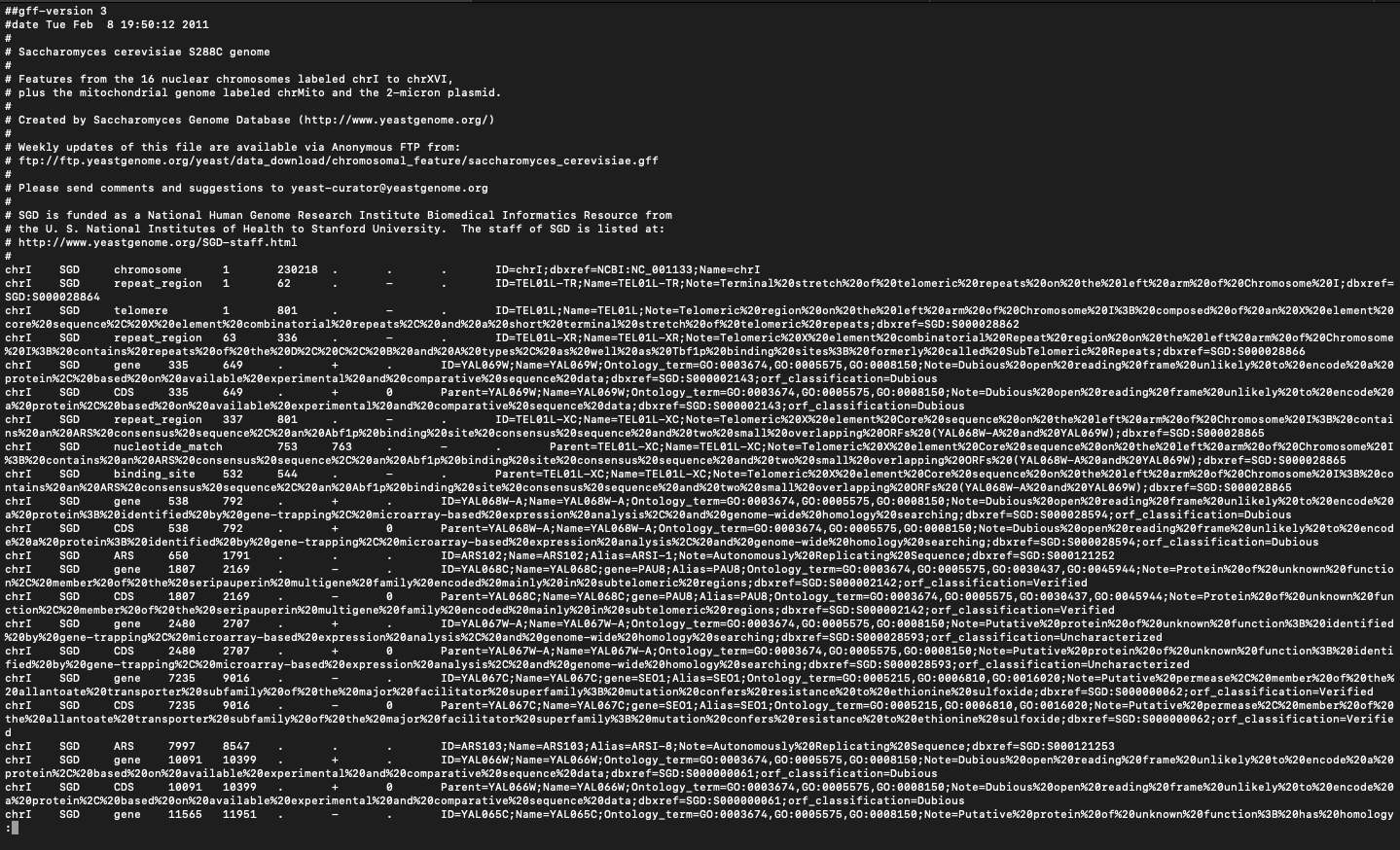
/home/jorvis1/Saccharomyces_cerevisiae_S288C.annotation.gff (**See attached images for sample**)
Note that this same file has both the annotation feature table and the FASTA sequence for the molecules referenced (See the '##FASTA' directive in the specification.)
Within the feature table, another column of note is the 9th, where we can store any key=value pairs relevant to that row's feature such as ID, Ontology_term, or Note.
Your task is to write a GFF3 feature exporter. A user should be able to run your script like this:
$ export_gff3_feature.py -source_gff=/path/to/some.gff3 -type=gene -attribute=ID ---value=YAR003
There are 4 arguments here that correspond to values in the GFF3 columns. In this case, your script should read the path to a GFF3 file, find any gene (column 3) which has an ID=YAR003W (column 9). When it finds this, it should use the coordinates for that feature (columns 4,5, and 7) and the FASTA sequence at the end of the document to return its FASTA sequence.
Your script should work regardless of the parameter values used, warning the user if no features were found that matched their query. (It should also check and warn if more than one feature matches the query.)
The output should just be printed on STDOUT (no writing to a file is necessary). It should have a header which matches their query, like this:
>gene:ID:YAR003W
... sequence here ...
Some bonus points will be awarded if you format the sequence portion of the FASTA output as 60-characters per line, which follows the standard.
Provide the complete source code AND the output of the program as it runs. You should do test runs with 3 features that are present in the file and 1 where you intentionally enter a feature NOT present in the file. Your script should handle this gracefully.
***Sample attachments of the GFF3 file provided. I also have the file for upload. Please provide an option for sending this file. Thanks.***
##gff-version 3 #date Tue Feb 8 19:50:12 2011 Saccharomyces cerevisiae $288C genome Features from the 16 nuclear chromosomes labeled chrI to chrXVI, plus the mitochondrial genome labeled chrMito and the 2-micron plasmid. Created by Saccharomyces Genome Database (http://www. yeastgenome. org/ ) Weekly updates of this file are available via Anonymous FTP from: ftp://ftp. yeastgenome . org/yeast/data_download/chromosomal_feature/saccharomyces_cerevisiae . off Please send comments and suggestions to yeast-curator@yeastgenome . org SGD is funded as a National Human Genome Research Institute Biomedical Informatics Resource from the U. S. National Institutes of Health to Stanford University. The staff of SGD is listed at: # http://www. yeastgenome . org/SGD-staff. html chri SGD chromosome 230218 ID=chrI; dbxref=NCBI : NC_001133; Name=chrI chrI repeat_region 62 ID=TELO1L-TR; Name=TELO1L-TR; Note=Terminal%20stretch%20of%%20telomeric%20repeats%20on%20the%20left%20arm*20of%20Chromosome%201 ; dbxref= SGD SGD : S000028864 chrI SGD telomere 1 801 ID=TELO1L; Name=TELO1L; Note=Telomeric%20region%20on%20the%20left%20arm%20of%20Chromosome%201%38%20composed%20of%20an%20*%20element%20 core%20sequence%20%20*%20element%20combinatorial%20repeats%20%20and%20%20short%20terminal%20stretch*20of%20telomeric%20repeats; dbxref=SGD : 5000028862 chrI SGD repeat_region 63 336 ID=TELOIL-XR; Name=TELOIL-XR; Note=Telomeric%20Xx20element%20combinatorial%20Repeat*20region%20on%20the%20left%20arm*20of%20Chromosome %20 1%38%20contains%20repeats%20of%20the%200%20%20C%20%208%20and%20A%20types%20%20as%20we11%20as%20Tbf1p%20binding%20sites%38%20formerly%20called%20SubTelomeric%20Repeats; dbxref=SGD: $060028866 chri SGD gene 335 649 ID=YAL069W; Name=YAL069W; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Dubious%20open%20reading%20frame%20unlikely%20to%20encode%20a%20 protein*20%20based%20on%20available%20experimental%20and%20comparative%20sequence%20data; dbxref=SGD: 5009002143; orf_classification=Dubious chri SGD CDS 335 649 Parent=YAL069W; Name=YAL069W; Ontology_term=GO: 6003674, GO: 6005575, GO: 0608150; Note=Dubious*20open%20reading%%20frame%20unlikely%20to%20encode%20 1%20protein%20%20based%20on%20available%20experimental%20and%20comparative%20sequence%20data; dbxref=SGD: S000002143; orf_classification=Dubious ID=TELOIL-XC; Name=TELOIL-XC; Note=Telomeric%20%20element%20Core%20sequence%20on%20the%20left%20arm%20of%20Chromosome%20 1%38%20contai chri SGD repeat_region 337 801 ns*20an*20ARS%20consensus%20sequence%20%20an%20Abf1p%20binding%20site%20consensus%20sequence%20and%20two%20small%20overlapping%200RF s%20 (YAL068W-A%20and%20YAL069W) ; dbxref=SGD : S000028865 chri SGD nucleotide_match 753 763 Parent=TEL01L-XC; Name=TEL01L-XC; Note=Telomeric%20X%20element%20Core%20sequence%20on%20the%20left%20arm%20of%20Chromosome%201 38%20contains%20an*20ARS%20consensus%20sequence%20%20an%20Abf1p%20binding%20site%20consensus%20sequence%20and%20two%20small%20overlapping%200RFs%20 (YAL068W-A%20and%20YAL069W) ; dbxref=SGD : S000028865 chrI SGD binding_site 532 544 Parent=TELOIL-XC; Name=TELOIL-XC; Note=Telomeric%20%20element%20Core%20sequence%20on%20the%20left%20arm%20of%20Chromosome%%20 1%%38%%20co ntains%20an%20ARS%20consensus%20sequence%20%20an%20Abf1p%20binding%20site%20consensus%20sequence%%20and%20two%20small%20overlapping%200RFs%20 (YAL068W-A%20and%%20YAL069W) ; dbxref=SGD: S000028865 gene ID=YAL068W-A; Name=YAL068W-A; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Dubious%20open%20reading%20frame%20unlikely%20to%20encode%20 chrI SGD 538 792 a*20protein*38%20identified%20by%20gene-trapping%20%20microarray-based%20expression%20analysis%20%20and%20genome-wide%20homology%20searching; dbxref=SGD: S009028594; orf_classification=Dubious chrI SGD CDS 538 792 Parent=YAL068W-A; Name=YAL068W-A; Ontology_term=GO: 0003674, GO: 0805575, GO: 0008150; Note=Dubious%20open%20reading%20frame%20unlikely%20to%20encod ex20a%20protein*38%20identified%20by%20gene-trapping%20%20microarray-based%20expression*20analysis%20%%20and%%20genome-wide%20homology%20searching; dbxref=SGD: $606028594; orf_classification=Dubious chri SGD ARS 650 1791 ID=ARS102; Name=ARS102; Alias=ARSI-1; Note=Autonomously%20Replicating*20Sequence; dbxref=SGD: 5000121252 180 2169 ID=YAL068C; Name=YAL068C; gene=PAUS; Alias=PAUS; Ontology_term=GO: 0003674, GO: 0005575, GO: 0030437 , GO: 0045944; Note=Protein%20of%20unknown%20functio chri SGD gene n%20%20member%20of%%20the%20seripauperin%20multigene%20family%20encoded%20mainly%20in%20subtelomeric%20regions ; dbxref=SGD: S090902142; orf_classification=Verified chri SGD CDS 1807 2169 Parent=YAL068C; Name=YAL068C; gene=PAUS; Alias=PAUS; Ontology_term=GO: 0003674, GO: 0005575, GO: 0030437, GO: 0045944; Note=Protein%20of%20unknown%20fun ction%20%20member%20of%20the%20seripauperin%20multigene%20family%20encoded%20mainly%20in%20subtelomeric%20regions; dbxref=SGD: S000002142; orf_classification=Verified ChrI SGD gene 2480 2707 ID=YAL067W-A; Name=YAL067W-A; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Putative%20protein%20of%20unknown%20function*38%20identified *20by%20gene-trapping%20%%20microarray-based%20expression%20analysis%20%20and%20genome-wide%20homology%20searching; dbxref=SGD: S000028593; orf_classification=Uncharacterized chri SGD CDS 2480 2707 Parent=YAL067W-A; Name=YAL067W-A; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Putative%20protein%20of%20unknown%20function%38%20identi fied%20by%20gene-trapping%20%20microarray-based%20expression%20analysis*20%20and%%20genome-wide%20homology%%20searching; dbxref=SGD: 5000028593; orf_classification=Uncharacterized SGD 7235 9016 ID=YAL067C; Name=YAL067C; gene=SEO1; Alias=SEO1; Ontology_term=GO: 0005215, GO: 0006810, GO: 0016020; Note=Putative*20permease%20%20member%20of%20the% chri gene 20allantoate%20transporter%20subfamily%20of%20the%20major%20facilitator%20superfamily*38%20mutation%20confers%20resistance%20to%20ethionine%20sulfoxide; doxref=SGD: 5000000062; orf_classification=Verified chri SGD CDS 7235 9016 Parent=YAL067C; Name=YAL067C; gene=SEO1; Alias=SEO1; Ontology_term=GO: 0005215, GO: 0006810, GO: 0016020; Note=Putative%20permease%20%%20member%20of%20 the%20allantoate%20transporter%20subfamily%20of%20the%20major%20facilitator%20superfamily%38%20mutation%20confers%20resistance%20to%20ethionine%20sulfoxide; dbxref=SGD: 5000000062; orf_classification=Verifie d chri SGD ARS 7997 8547 ID=ARS103; Name=ARS103; Alias=ARSI-8; Note=Autonomously%20Replicating%20Sequence; dbxref=SGD: 5000121253 10091 10399 ID=YAL066W; Name=YAL066W; Ontology_term=GO: 0603674, GO: 0605575, GO: 0608150; Note=Dubious%20open%20reading%20frame%20unlikely%20to%20encode%20a%20 chrI SGD gene protein*20%20based%20on%20available%20experimental%20and%20comparative%20sequence%20data ; dbxref=SGD : 5000000061; orf_classification=Dubious Parent=YAL066W; Name=YAL066W; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Dubious%20open%20reading%20frame%20unlikely%20to%20encode%20 chrI SGD CDS 10091 10399 2%20protein%20%20based%20on%20available%20experimental%20and%20comparative%20sequence%20data; dbxref=SGD: S000000061; orf_classification=Dubious chrI SGD gene 11565 11951 ID=YAL065C; Name=YAL065C; Ontology_term=GO: 0003674, GO: 0005575, GO: 0008150; Note=Putative%20protein%20of%20unknown%20function%38%20has%20homology##FASTA >chri CCACACCACACCCACACACCCACACACCACACCACACACCACACCACACCCACACACACACATCCTAACACTACCCTAAC ACAGCCCTAATCTAACCCTGGCCAACCTGTCTCTCAACTTACCCTCCATTACCCTGCCTCCACTCGTTACCCTGTCCCAT TCAACCATACCACTCCGAACCACCATCCATCCCTCTACTTACTACCACTCACCCACCGTTACCCTCCAATTACCCATATC CAACCCACTGCCACTTACCCTACCATTACCCTACCATCCACCATGACCTACTCACCATACTGTTCTTCTACCCACCATAT TGAAACGCTAACAAATGATCGTAAATAACACACACGTGCTTACCCTACCACTTTATACCACCACCACATGCCATACTCAC CCTCACTTGTATACTGATTTTACGTACGCACACGGATGCTACAGTATATACCATCTCAAACTTACCCTACTCTCAGATTC CACTTCACTCCATGGCCCATCTCTCACTGAATCAGTACCAAATGCACTCACATCATTATGCACGGCACTTGCCTCAGCGG TCTATACCCTGTGCCATTTACCCATAACGCCCATCATTATCCACATTTTGATATCTATATCTCATTCGGCGGTCCCAAAT ATTGTATAACTGCCCTTAATACATACGTTATACCACTTTTGCACCATATACTTACCACTCCATTTATATACACTTATGTC AATATTACAGAAAAATCCCCACAAAAATCACCTAAACATAAAAATATTCTACTTTTCAACAATAATACATAAACATATTG GCTTGTGGTAGCAACACTATCATGGTATCACTAACGTAAAAGTTCCTCAATATTGCAATTTGCTTGAACGGATGCTATTT CAGAATATTTCGTACTTACACAGGCCATACATTAGAATAATATGTCACATCACTGTCGTAACACTCTTTATTCACCGAGC AATAATACGGTAGTGGCTCAAACTCATGCGGGTGCTATGATACAATTATATCTTATTTCCATTCCCATATGCTAACCGCA ATATCCTAAAAGCATAACTGATGCATCTTTAATCTTGTATGTGACACTACTCATACGAAGGGACTATATCTAGTCAAGAC GATACTGTGATAGGTACGTTATTTAATAGGATCTATAACGAAATGTCAAATAATTTTACGGTAATATAACTTATCAGCGG CGTATACTAAAACGGACGTTACGATATTGTCTCACTTCATCTTACCACCCTCTATCTTATTGCTGATAGAACACTAACCC CTCAGCTTTATTTCTAGTTACAGTTACACAAAAAACTATGCCAACCCAGAAATCTTGATATTTTACGTGTCAAAAAATGA GGGTCTCTAAATGAGAGTTTGGTACCATGACTTGTAACTCGCACTGCCCTGATCTGCAATCTTGTTCTTAGAAGTGACGC ATATTCTATACGGCCCGACGCGACGCGCCAAAAAATGAAAAACGAAGCAGCGACTCATTTTTATTTAAGGACAAAGGTTG CGAAGCCGCACATTTCCAATTTCATTGTTGTTTATTGGACATACACTGTTAGCTTTATTACCGTCCACGTTTTTTCTACA ATAGTGTAGAAGTTTCTTTCTTATGTTCATCGTATTCATAAAATGCTTCACGAACACCGTCATTGATCAAATAGGTCTAT AATATTAATATACATTTATATAATCTACGGTATTTATATCATCAAAAAAAAGTAGTTTTTTTATTTTATTTTGTTCGTTA ATTTTCAATTTCTATGGAAACCCGTTCGTAAAATTGGCGTTTGTCTCTAGTTTGCGATAGTGTAGATACCGTCCTTGGAT AGAGCACTGGAGATGGCTGGCTTTAATCTGCTGGAGTACCATGGAACACCGGTGATCATTCTGGTCACTTGGTCTGGAGC AATACCGGTCAACATGGTGGTGAAGTCACCGTAGTTGAAAACGGCTTCAGCAACTTCGACTGGGTAGGTTTCAGTTGGGT GGGCGGCTTGGAACATGTAGTATTGGGCTAAGTGAGCTCTGATATCAGAGACGTAGACACCCAATTCCACCAAGTTGACT CTTTCGTCAGATTGAGCTAGAGTGGTGGTTGCAGAAGCAGTAGCAGCGATGGCAGCGACACCAGCGGCGATTGAAGTTAA TTTGACCATTGTATTTGTTTTGTTTGTTAGTGCTGATATAAGCTTAACAGGAAAGGAAAGAATAAAGACATATTCTCAAA GGCATATAGTTGAAGCAGCTCTATTTATACCCATTCCCTCATGGGTTGTTGCTATTTAAACGATCGCTGACTGGCACCAG TTCCTCATCAAATATTCTCTATATCTCATCTTTCACACAATCTCATTATCTCTATGGAGATGCTCTTGTTTCTGAACGAA TCATAAATCTTTCATAGGTTTCGTATGTGGAGTACTGTTTTATGGCGCTTATGTGTATTCGTATGCGCAGAATGTGGGAA TGCCAATTATAGGGGTGCCGAGGTGCCTTATAAAACCCTTTTCTGTGCCTGTGACATTTCCTTTTTCGGTCAAAAAGAAT ATCCGAATTTTAGATTTGGACCCTCGTACAGAAGCTTATTGTCTAAGCCTGAATTCAGTCTGCTTTAAACGGCTTCCGCG GAGGAAATATTTCCATCTCTTGAATTCGTACAACATTAAACGTGTGTTGGGAGTCGTATACTGTTAGGGTCTGTAAACTT GTGAACTCTCGGCAAATGCCTTGGTGCAATTACGTAATTTTAGCCGCTGAGAAGCGGATGGTAATGAGACAAGTTGATAT CAAACAGATACATATTTAAAAGAGGGTACCGCTAATTTAGCAGGGCAGTATTATTGTAGTTTGATATGTACGGCTAACTG AACCTAAGTAGGGATATGAGAGTAAGAACGTTCGGCTACTCTTCTTTCTAAGTGGGATTTTTCTTAATCCTTGGATTCTT AAAAGGTTATTAAAGTTCCGCACAAAGAACGCTTGGAAATCGCATTCATCAAAGAACAACTCTTCGTTTTCCAAACAATC TTCCCGAAAAAGTAGCCGTTCATTTCCCTTCCGATTTCATTCCTAGACTGCCAAATTTTTCTTGCTCATTTATAATGATT GATAAGAATTGTATTTGTGTCCCATTCTCGTAGATAAAATTCTTGGATGTTAAAAAATTATTATTTTCTTCATAAAGAAG CTTTCAAGATATAAGATACGAAATAGGGGTTGATAATTGCATGACAGTAGCTTTAGATCAAAAAGGAAAGCATGGAGGGA AACAGTAAACAGTGAAAATTCTCTTGAGAACCAAAGTAAACCTTCATTGAAGAGCTTCCTTAAAAAATTTAGAATCTCCC ATGTCAACGGGTTTCCATACCTCCCCAGCATCATACATCTTTTTTCAAAGAAACTTCAAATGCCTCTTTTATGCAAGGGG CAAAATCCTGAAATGACTTAAACTTAGCAGTTTCGTCTTTTTTCAAAGAGAATGGTTGAAGAAGAATTGTTTTGGACGCT TATTGACAATCTGTTGCATTGATAAAGTACCTACTATCCCAGACTATATTTGTATACAAGTACAAAATTAGGTTTGTTGA AACAACTTTCCGATCATTGGTGCCCGTATCTGATGTTTTTTTAGTAATTTCTTTGTAAATACAGGGAGTTGTTTCGAAAG CTTATGAGAAAAATACATGAATGACAGGTAAAAATATTGGCTCGAAAAAGAGGACAAAAAGAGAAATCATAAATGAGTAA ACCCACTTGCTGGACATTATCCAGTAAAGGCTTGGTAGTAACCATAATATTACCCAGGTACGAAACGCTAAGAACTTGAA AGACTCATAAAACTTCCAGGTTAAGCTATTTTTGAAAATATTCTGAGGTAAAAGCCATTAAGGTCCAGATAACCAAGGGA CAATAAACCTATGCTTTTCTTGTCTTCAATTTCAGTATCTTTCCATTTTGATAATGAGCTAGTGATCCGGAAAGCTACTT TATGATGTTTCAAGGCCTGAAGTTTGAATATTTATGTAGTTCAACATCAAATGTGTCTATTTTGTGATGAGGCAACCGTC GACAACCTTATTATCGAAAAAGAACAACAAGTTCACATGCTTGTTACTCTCTATAACTAGAGAGTACTTTTTTTGGAAGC AAGTAAGAATAAGTCAATTTCTACTTACCTCATTAGGGAAAAATTTAATAGCAGTTGTTATAACGACAAATACAGGCCCT AAAAAATTCACTGTATTCAATGGTCTACGAATCGTCAATCGCTTGCGGTTATGGCACGAAGAACAATGCAATAGCTCTTA CAAGCCACTACATGACAAGCAACTCATAATTTAAGTGGATAGCTTGTGATAAATTGAATTTTCTCTGTTTAGTACTTGCC GAATAGTTACTTGTTAGTTGCAGATGCTTTTTGATGACAAAGTTATCAATCTCAATATTAA TAGGCTTTCAGGT