Question
There is the code. from PIL import Image import numpy as np import matplotlib.pyplot as plt from scipy import signal import math from demos.demo02 import
 There is the code.
There is the code.
from PIL import Image import numpy as np import matplotlib.pyplot as plt from scipy import signal import math from demos.demo02 import harris
def plot_1D_gaussian(mu, variance, color='orange', x_limit=3): sigma = math.sqrt(variance) # get 100 'x' values between mu - x_limit * sigma and mu + x_limit * sigma x = np.linspace(mu - x_limit * sigma, mu + x_limit * sigma, 100) y = np.exp(-np.power(x - mu, 2.) / (2 * variance))/(np.sqrt(2 * np.pi * variance)) plt.plot(x, y, color)
def plot_2D_surface(matrix_2d): kernel_size = matrix_2d.shape X, Y = np.meshgrid(np.linspace(-kernel_size[0] // 2, kernel_size[0] // 2, kernel_size[0]), np.linspace(-kernel_size[1] // 2, kernel_size[1] // 2, kernel_size[1])) plt.figure() ax = plt.axes(projection='3d') ax.plot_surface(X, Y, matrix_2d)
def gaussian_2D_kernel(size=5, sigma=3.0): # get x, y coordinates using np.meshgrid function x, y = np.meshgrid(np.linspace(-1, 1, size), np.linspace(-1, 1, size)) # YOUR CODES #1 6 points # Step 1: Compute a 2D gaussian kernel and store it in kernel # Step 2: Compute the normalized kernel and store it in kernel_norm # Hint: the sum of the numbers in the kernel is 1.0 kernel kernel_norm print("2D Gaussian-like array:") print(kernel_norm) return kernel_norm
def main(): path = '../../data/empire.jpg' im = np.array(Image.open(path).convert('L'))
""" Part 1 Gaussian """ kernel_size = 11 # odd number # get gaussian kernel kernel = gaussian_2D_kernel(size=kernel_size, sigma=2.0)
# Apply Gaussian Blurring to the image using convolution im_blurring = signal.convolve2d(im, kernel, boundary='symm', mode='same')
""" Part 2 Harris Corner Detector """ im_harris = harris.compute_harris_response(im) # Get harris points by setting min_dist = 30 and threshold = 0.01 # Store the results in a numpy array filtered_cords # YOUR CODES #2 2 points filtered_cords
""" Part 3 Homography """ # The corners of a rectangle are located at (-0.6, -0.3), (0.6, -0.3), (0.6, 0.3) and (-0.6, 0.3) # Use a two-row numpy array to store the coordinates # YOUR CODES #3 3 points points
# Use np.vstack to obtain the homogeneous coordinates # YOUR CODES #4 3 points points_norm
# Define two affine transformation matrices. # Obtain the new coordinates and store them in points_norm_s and points_norm_sr # YOUR CODES #5 9 points # Scaling
points_norm_s # Rotation (about the origin)
points_norm_sr
# Plot the Kernel plot_2D_surface(kernel)
# construct a second figure plt.figure() plt.gray() # use subplot to display the images using a 2 by 2 setting plt.subplot(2, 2, 1) plot_1D_gaussian(0, 0.2, 'b') # YOUR CODES #6 4 points
plt.subplot(2, 2, 2) # YOUR CODES #7 3 points # On the blurred image, show 10 strongest harris points using cyan marks
plt.subplot(2, 2, 3) plt.axis('equal') plt.plot(points_norm[0], points_norm[1], 'g')
plt.subplot(2, 2, 4) plt.axis('equal') plt.plot(points_norm_s[0], points_norm_s[1], 'b') plt.plot(points_norm_sr[0], points_norm_sr[1], 'r') plt.show()
if __name__ == '__main__': main()
YOUR ONLY NEED FINISH THE CODE 3 AND CODE 4
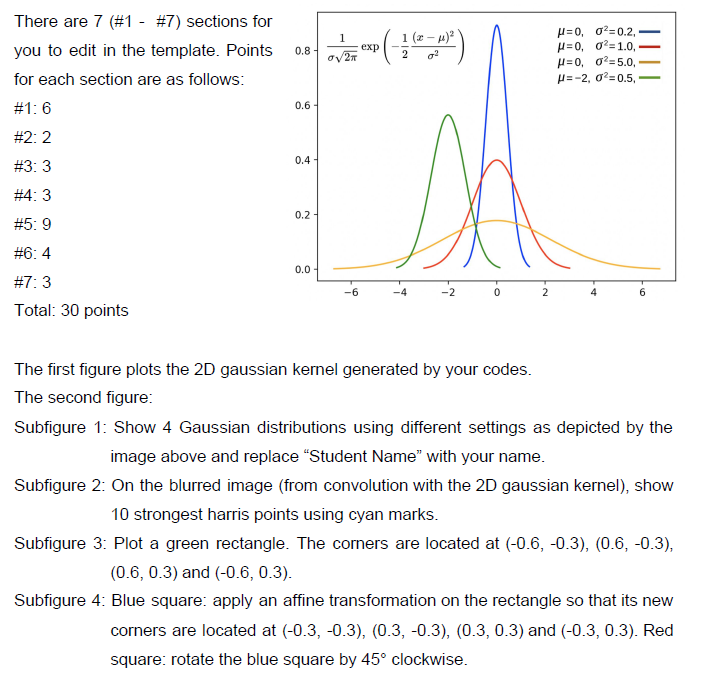
1 1 (2-4) g? 0.8 exp 2 There are 7 (#1 - #7) sections for you to edit in the template. Points for each section are as follows: #1: 6 027 =0,02=0.2, =0,02=1.0, p=0,02=5.0 = -2, 02-0.5, 0.6 #2:2 0.4 #3:3 #4:3 Da 0.2 #5: 9 #6:4 0.0 -6 -2 0 2 6 #7: 3 Total: 30 points The first figure plots the 2D gaussian kernel generated by your codes. The second figure: Subfigure 1: Show 4 Gaussian distributions using different settings as depicted by the image above and replace Student Name" with your name. Subfigure 2: On the blurred image (from convolution with the 2D gaussian kernel), show 10 strongest harris points using cyan marks. Subfigure 3. Plot a green rectangle. The corners are located at (-0.6, -0.3), (0.6, -0.3), (0.6, 0.3) and (-0.6, 0.3). Subfigure 4: Blue square: apply an affine transformation on the rectangle so that its new corners are located at (-0.3, -0.3), (0.3,-0.3), (0.3, 0.3) and (-0.3, 0.3). Red square: rotate the blue square by 45 clockwise. 1 1 (2-4) g? 0.8 exp 2 There are 7 (#1 - #7) sections for you to edit in the template. Points for each section are as follows: #1: 6 027 =0,02=0.2, =0,02=1.0, p=0,02=5.0 = -2, 02-0.5, 0.6 #2:2 0.4 #3:3 #4:3 Da 0.2 #5: 9 #6:4 0.0 -6 -2 0 2 6 #7: 3 Total: 30 points The first figure plots the 2D gaussian kernel generated by your codes. The second figure: Subfigure 1: Show 4 Gaussian distributions using different settings as depicted by the image above and replace Student Name" with your name. Subfigure 2: On the blurred image (from convolution with the 2D gaussian kernel), show 10 strongest harris points using cyan marks. Subfigure 3. Plot a green rectangle. The corners are located at (-0.6, -0.3), (0.6, -0.3), (0.6, 0.3) and (-0.6, 0.3). Subfigure 4: Blue square: apply an affine transformation on the rectangle so that its new corners are located at (-0.3, -0.3), (0.3,-0.3), (0.3, 0.3) and (-0.3, 0.3). Red square: rotate the blue square by 45 clockwiseStep by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


