Answered step by step
Verified Expert Solution
Question
1 Approved Answer
Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct

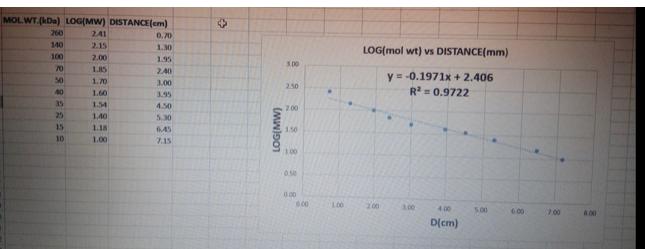

Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve Using the migration of the DNA markers (Invitrogen 1 Kb Plus DNA ladder) on the agarose gel with your PCR product you should construct a standard curve of the migration rate of the DNA markers against the log10 value of their molecular weight. The table below is provided to tabulate the data necessary for making this graph. NOTE: Only the middle column (Log10 size of DNA marker) will be graded. Do not enter any units (i.e. bp) in this column and give your answer to 2 significant figures. Molecular weight and migration of DNA markers Size of DNA marker (bp) 3000 2000 1500 1000 850 650 500 400 300 200 100 Logo size of DNA marker (bp) Distance migrated by DNA marker from top of agarose gel (mm) MOLWT.(kDa) LOG(MW) DISTANCE(cm) 241 2.15 2.00 1.85 1.70 1.60 1.54 1,40 1.18 1.00 260 140 100 553588888 70 50 40 25 10 0.70 1.30 1.95 2.40 3.00 3.95 4.50 5.30 6.45 7.15 + (MW)901 5.00 2.50 200 1.50 1.00 0.500 0.00 0.00 100 LOG(mol wt) vs DISTANCE(mm) y = -0.1971x + 2.406 R = 0.9722 200 4.00 D(cm) 5.00 6.00 7.00 400 Using your standard curve, calculate the observed size of the two PCR products generated from pOTC and pOTCA and express this base pairs (p) To do this, you will need to measure the migration of the two PCR products on the agerose gel and use the distance migrated to interpolate their logis molendar weght Compare the values you estimate from your standard curve to those you calculated previously in question 12 and provide one way you tright improve the source of the sizes you calculate a standard curve
Step by Step Solution
There are 3 Steps involved in it
Step: 1
To construct a standard curve for the Invitrogen 1 Kb Plus DNA ladder using the migration rates of t...
Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


