Question
vicsek2d matlab code: function gobs=vicsek2d(N, ts, eta, top, topval, gobsFlag, boundary, show) % function X=vicsek2d(N, ts, eta, rad, show) % % eta is the the
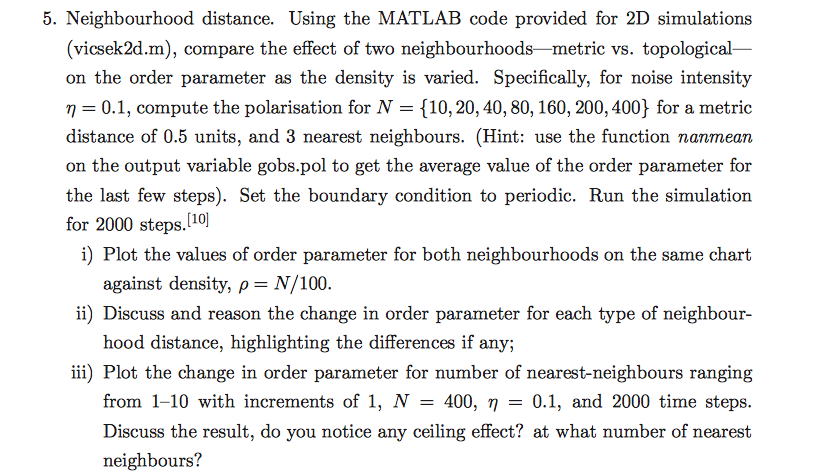
vicsek2d matlab code:
function gobs=vicsek2d(N, ts, eta, top, topval, gobsFlag, boundary, show)
% function X=vicsek2d(N, ts, eta, rad, show)
%
% eta is the the noise value for dtheta
% ts number of timesteps
% N is the number of birds
% top is the topology ('metric', 'nn')
% topval is the value for metric, radius, for nn number of nearest
% neighbors
% gobsFlag is 1 for computing global observables and 0 for not
% boundary is 0 for periodic and 1 for reflective
% show is 1 if you want to plot. set to 0 for analysis
%
% Vicsek, T., Czirak, A., Ben-Jacob, E., Cohen, I., & Shochet, O. (1995).
% Novel Type of Phase Transition in a System of Self-Driven Particles.
% Physical Review Letters, 75, 1226-1229. doi:10.1103/PhysRevLett.75.1226
% s = RandStream('mt19937ar','Seed',1);
% RandStream.setGlobalStream(s);
% default params if you run it as is
if nargin ==0
N=400; ts=500; eta=pi/6; top='metric'; topval=2; show=1;
gobsFlag=0;
boundary=2; % 0=periodic, 1=reflective, 2=circular
end
gobs.annd=zeros(1,ts);
gobs.pol=zeros(1,ts);
gobs.angm=zeros(1,ts);
gk=ts-min(ts/10, 40);
% note that we assign units but these could be anything to match whatever
% is realistic
b=[-5 5 -5 5]; % boundary (m)
sp=.03; % speed m/s
dt=1; % seconds?
% initialize
x1=zeros(N,ts); x2=x1; v1=x1; v2=x1;
% use component-wise to speed up code
x1(:,1)=b(1)+rand(N,1)*(b(2)-b(1)); % x_1
x2(:,1)=b(3)+rand(N,1)*(b(4)-b(3)); % x_2
if boundary==2
% note that this may not be the correct way to sample uniformly within
% a circle, but we will approx. since it should not affect our final
% results.
thk=rand(N,1)*2*pi;
x1(:,1)=(b(2)-b(1))/2*rand(N,1).*cos(thk);
x2(:,1)=(b(2)-b(1))/2*rand(N,1).*sin(thk);
end
v1(:,1)=randn(N,1); % xdot_1 / v_1 (direction only)
v2(:,1)=randn(N,1); % xdot_2 / v_2 (direction only)
for k=1:ts
% topology
D=sqrt((x1(:,k)*ones(1,N)-ones(N,1)*x1(:,k)').^2 + ...
(x2(:,k)*ones(1,N)-ones(N,1)*x2(:,k)').^2);
% motion model | equation (1) in paper
x1(:,k+1)=x1(:,k)+sp*v1(:,k)*dt;
x2(:,k+1)=x2(:,k)+sp*v2(:,k)*dt;
for ii=1:N
% interaction rule
% metric
if strcmp(top, 'metric')
rad=topval;
nidx=find(D(ii,:)
elseif strcmp(top, 'nn')
% or n nearest neighbors
nn=topval;
[~, nidx]=sort(D(ii,:));
nidx=nidx(1:nn); % including ii
end
% average angle
sin_th=sum(v2(nidx,k))umel(nidx);
cos_th=sum(v1(nidx,k))umel(nidx);
if ~isempty(nidx)
th_hat_ri=atan2(sin_th, cos_th);
else
th_hat_ri=0;
end
% direction update | equation (2) in paper
th=th_hat_ri-eta/2 + rand*eta;
% update the direction for the next time-step
v1(ii,k+1)=cos(th);
v2(ii,k+1)=sin(th);
end
if boundary==0
% periodic boundary condition
x1(x1(:,k+1)
x1(x1(:,k+1)>b(2),k+1)=x1(x1(:,k+1)>b(2),k+1)-(b(2)-b(1));
x2(x2(:,k+1)
x2(x2(:,k+1)>b(4),k+1)=x2(x2(:,k+1)>b(4),k+1)-(b(4)-b(3));
elseif boundary==1
% reflective boundary condition
v1(x1(:,k+1)
x1(x1(:,k+1)
v1(x1(:,k+1)>b(2),k+1)=-v1(x1(:,k+1)>b(2),k+1);
x1(x1(:,k+1)>b(2),k+1)=x1(x1(:,k+1)>b(2),k+1)+v1(x1(:,k+1)>b(2),k+1)*sp;
v2(x2(:,k+1)
x2(x2(:,k+1)
v2(x2(:,k+1)>b(4),k+1)=-v2(x2(:,k+1)>b(4),k+1);
x2(x2(:,k+1)>b(4),k+1)=x2(x2(:,k+1)>b(4),k+1)+v2(x2(:,k+1)>b(4),k+1)*sp;
elseif boundary==2
% reflective circular boundary
rk=sqrt(x1(:,k+1).^2+x2(:,k+1).^2);
ct=x1(:,k+1)./rk;
st=x2(:,k+1)./rk;
outidx=(rk>(b(2)-b(1))/2);
if sum(outidx)
% v=vt+vn
% v'=vt-vn=vt+vn-2*vn=v-2*vn;
% vn1=x1*v1/rk
vn=v1(:,k+1).*ct+v2(:,k+1).*st;
v1(outidx,k+1)=v1(outidx,k+1)-2*ct(outidx).*vn(outidx);
v2(outidx,k+1)=v2(outidx,k+1)-2*st(outidx).*vn(outidx);
nv=sqrt(v1(outidx,k+1).^2+v2(outidx,k+1).^2);
v1(outidx,k+1)=v1(outidx,k+1).v;
v2(outidx,k+1)=v2(outidx,k+1).v;
x1(outidx, k+1)=x1(outidx, k+1)+v1(outidx,k+1)*sp;
x2(outidx, k+1)=x2(outidx, k+1)+v2(outidx,k+1)*sp;
end
end
if gobsFlag
% only compute for the last tenth of the time
if k > gk
gobs.annd(1,k)=annd(D);
gobs.pol(1,k)=pol([v1(:,k)'; v2(:,k)']);
gobs.angm(1,k)=ang_m([v1(:,k)'; v2(:,k)'], [x1(:,k)'; x2(:,k)']);
else
gobs.annd(1,k)=nan;
gobs.pol(1,k)=nan;
gobs.angm(1,k)=nan;
end
end
% display; alternatively use the output for analysis
if show
figure(1); gcf;
if gobsFlag
subplot(2,2,1); gca; cla;
else
clf;
end
plot(x1(:,k), x2(:,k), 'b.');
hold on;
axis(b);
axis square;
quiver(x1(:,k), x2(:,k), v1(:,k)*.25, v2(:,k)*.25, 0, 'k');
xlabel(sprintf('[%d]', k));
if boundary==2
th=linspace(-pi, pi, 50);
plot((b(2)-b(1))/2*cos(th), (b(2)-b(1))/2*sin(th), 'b');
end
if gobsFlag
% annd
subplot(2,2,2); gca; cla;
plot(1:k, gobs.annd(1,1:k), 'linewidth', 2);
hold on;
xlabel('time'); ylabel('annd (m)');
axis([0 ts 0 (b(2)-b(1))/2]);
% pol
subplot(2,2,3); gca; cla;
plot(1:k, gobs.pol(1,1:k), 'linewidth', 2);
hold on;
xlabel('time'); ylabel('pol');
axis([0 ts 0 1]);
% angular momentum
subplot(2,2,4); gca; cla;
plot(1:k, gobs.angm(1,1:k), 'linewidth', 2);
hold on;
xlabel('time'); ylabel('normalized angular momentum');
axis([0 ts 0 1]);
end
drawnow;
end
end
function [dat, perm]=annd(D)
% function annd(r)
% average nearest-neighbor distance
%
% r is a d x n matrix with d dimensions and n targets
D=D+eye(size(D,1))*100000000;
[perm, ~]=min(D);
dat=mean(perm);
function P=pol(v)
% function pol(v)
%
% v is a d x n matrix with d dimensions and n targets
[d, n]=size(v);
vhat = v./(ones(d,1)*sqrt(sum(v.^2)));
P=1*norm(sum(vhat,2));
function am=ang_m(v,rf)
[d, n]=size(v);
vhat = v./(ones(d,1)*sqrt(sum(v.^2)));
rf=rf-mean(rf,2)*ones(1,size(rf,2));
if size(rf,2) == size(v,2)
rf=[rf; zeros(1,size(rf,2))];
vhat=[vhat; zeros(1,size(vhat,2))];
am=norm(sum(cross(rf,vhat),2))/sum(sqrt(sum(rf.^2)).*sqrt(sum(vhat.^2)));
% am=dot(sum(cross(rf,vhat),2), [0 0 1]')/sum(sqrt(sum(cross(rf,vhat).^2,1)));
% am=dot(sum(cross(rf,vhat),2), [0 0 1]')/sum(sqrt(sum(rf.^2)).*sqrt(sum(vhat.^2)));
% am=norm(dot(sum(cross(rf,vhat),2), [0 0 1]'));
% am=dot(sum(cross(rf,vhat),2), [0 0 1]');
% am=norm(dot(sum(cross(rf,vhat),2), [0 0 1]'));
else
am=nan;
end
5. Neighbourhood distance. Using the MATLAB code provided for 2D simulations (vicsek2d.m), compare the effect of two neighbourhoods metric vs. topological on the order parameter as the density is varied. Specifically, for noise intensity = 0.1, compute the polarisation for N = {10, 20, 40, 80, 160, 200, 400} for a metric distance of 0.5 units, and 3 nearest neighbours. (Hint: use the function nanmean on the output variable gobs.pol to get the average value of the order parameter for the last few steps). Set the boundary condition to periodic. Run the simulation for 2000 steps. [I i) Plot the values of order parameter for both neighbourhoods on the same chart ii) Discuss and reason the change in order parameter for each type of neighbour- ) Plot the change in order parameter for number of nearest-neighbours ranging 400, = 0.1, and 2000 time steps. Discuss the result, do you notice any ceiling effect? at what number of nearest hood distance, highlighting the differences if any; from 1-10 with increments of 1, N neighbours? 5. Neighbourhood distance. Using the MATLAB code provided for 2D simulations (vicsek2d.m), compare the effect of two neighbourhoods metric vs. topological on the order parameter as the density is varied. Specifically, for noise intensity = 0.1, compute the polarisation for N = {10, 20, 40, 80, 160, 200, 400} for a metric distance of 0.5 units, and 3 nearest neighbours. (Hint: use the function nanmean on the output variable gobs.pol to get the average value of the order parameter for the last few steps). Set the boundary condition to periodic. Run the simulation for 2000 steps. [I i) Plot the values of order parameter for both neighbourhoods on the same chart ii) Discuss and reason the change in order parameter for each type of neighbour- ) Plot the change in order parameter for number of nearest-neighbours ranging 400, = 0.1, and 2000 time steps. Discuss the result, do you notice any ceiling effect? at what number of nearest hood distance, highlighting the differences if any; from 1-10 with increments of 1, N neighboursStep by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


