Question
With the following matlab code, and the filepath being a txt file of RNAseq gene expression data of 500 cancer patients (http://m.uploadedit.com/bbtc/1510541260973.txt) : Using the
With the following matlab code, and the filepath being a txt file of RNAseq gene expression data of 500 cancer patients (http://m.uploadedit.com/bbtc/1510541260973.txt) :
Using the randi function (if you use MATLAB) select randomly 2,000 genes out of 18,026 and 50 patients out of 500 patients in this dataset (so now you have a matrix of 2,000 by 50) and perform the following clustering approaches 1- use Kmeans clustering and cluster these genes into 20 clusters--- note you cluster genes and not patients. Plot the centers of each cluster and report the number of genes in each cluster. Do you think you need to increase or decrease the number of cluster in order to have a better clustering for these data. 2- do the same using hierarchical clustering and display dendrogram and heat map of this clustering.
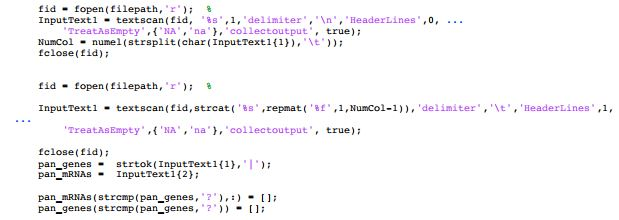
fid fopen (filepath, r InputTextl -textsean (fid, %s. ,1,' delimiter' ,' ","HeaderLines. ,0, TreatAsEmpty,NA,na', "collectoutput,true) Numcolnumel(strsplit (char InputTextl1)),)) fclose(fid) fid- fopen (filepath,'r'); % InputText1 textscan (fid, streat ( '%s' ,repmat ( 'BE, ,1,NumCol-1)),'delimiter','\t','HeaderLines' ,1, - TreatAsEmpty' , { ' NA. , na'),' collectoutput, , true) ; felose(fid pan-genes strtok (InputText1(1),'-) ; pan mRNAs- InputText1 (2) pan mRNAs (stremp (pan genes,, pan-genes ( stremp (pan-genes,-?)) - [ ] ; fid fopen (filepath, r InputTextl -textsean (fid, %s. ,1,' delimiter' ,' ","HeaderLines. ,0, TreatAsEmpty,NA,na', "collectoutput,true) Numcolnumel(strsplit (char InputTextl1)),)) fclose(fid) fid- fopen (filepath,'r'); % InputText1 textscan (fid, streat ( '%s' ,repmat ( 'BE, ,1,NumCol-1)),'delimiter','\t','HeaderLines' ,1, - TreatAsEmpty' , { ' NA. , na'),' collectoutput, , true) ; felose(fid pan-genes strtok (InputText1(1),'-) ; pan mRNAs- InputText1 (2) pan mRNAs (stremp (pan genes,, pan-genes ( stremp (pan-genes,-?)) - [ ]Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


