Write another method for the Alignment class called getConsensus. It should construct and return a new Sequence from the consensus characters for each column in an alignment. You are recommended to repeatedly call getConsensusForColumn in your implementation. Note that the returned sequence should be composed from the same alphabet as the alignment. Use your `getConsensus` to provide the consensus sequence for the five sequences above
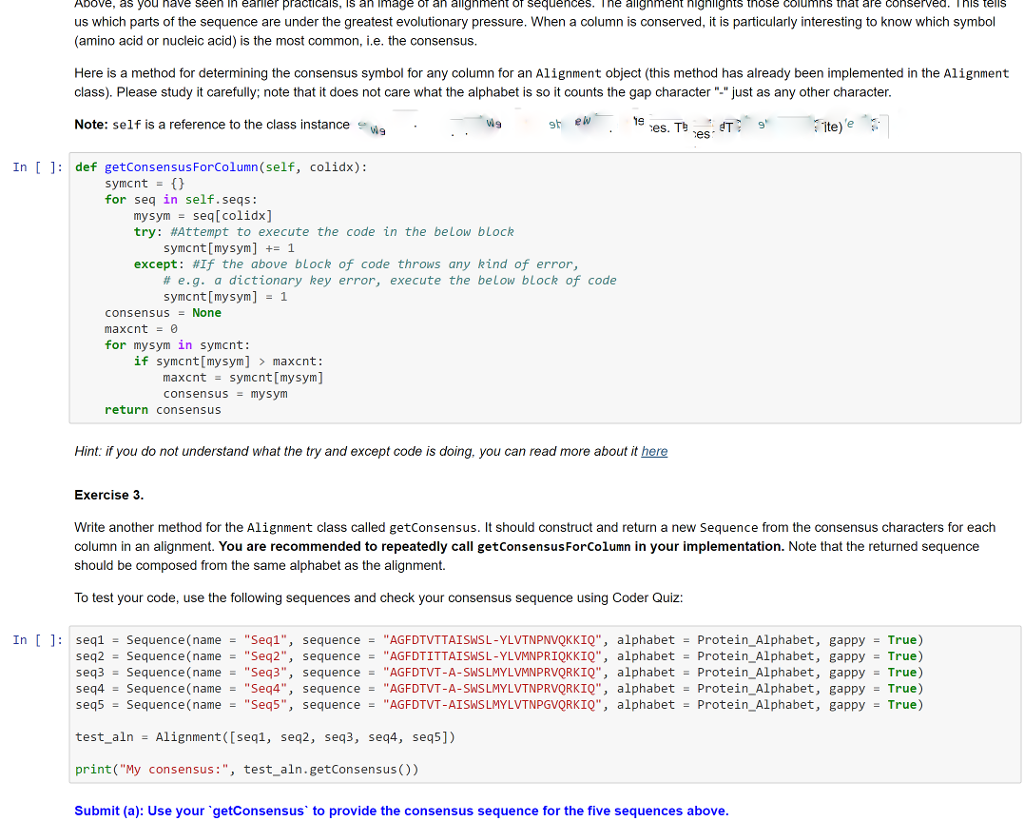
Above, as you nave seen in eaner practicals, s an image an alignment OT sequences. | ne alignment nignlignts tnose columns that are conserved. nis tells us which parts of the sequence are under the greatest evolutionary pressure. When a column is conserved, it is particularly interesting to know which symbol (amino acid or nucleic acid) is the most common, .e. the consensus. Here is a method for determining the consensus symbol for any column for an Alignment object (this method has already been implemented in the Alignment class). Please study it carefully; note that it does not care what the alphabet is so it counts the gap character ".just as any other character Note: self is a reference to the class instance 19 es. T es ite)e In [ 1: def getConsensusForColumn(self, colidx): for seq in self.seqs: mysym - seq[colidx] try: #Attempt to execute the code in the below block symcnt [mysym] 1 except: #If the above block of code throws any kind of error, # e.g. a dictionary key error, execute the below block of code symcnt [mysym]1 consensus None maxcnt0 for mysym in symcnt: if symcnt[mysym] maxcnt: maxcntsymcnt [mysym] consensus - mysym return consensus Hint: if you do not understand what the try and except code is doing, you can read more about it here Exercise 3 Write another method for the Alignment class called getConsensus. It should construct and return a neW Sequence from the consensus characters for each column in an alignment. You are recommended to repeatedly call getConsensusForColumn in your implementation. Note that the returned sequence should be composed from the same alphabet as the alignment. To test your code, use the following sequences and check your consensus sequence using Coder Quiz: In [ ] : = Sequence(name- "Seq1", sequence- "AGFDTVTTA?SWSL-YLVTNPNVOKKIO", alphabet = Sequence(name- "Seq2", sequence = "AGFDTITTAISWSL-YLVMNPR1QKK10", alphabet Sequence(name = "Seq3", sequence = "AGFDTVT-A-SWS LMYLVMNPRVORK10", alphabet = Sequence(name = "Seq4", sequence- "AGFDTVT-A-SWSLMYLVTNPRVORK10", alphabet Sequence (name "Seq5", sequence"AGFDTVT-AISWSLMYLVTNPGVQRKIQ", alphabet Protein-Alphabet, seq1 seq2 seq3 seq4 seq5 - True) Protein-Alphabet, gappy - True) Protein. Alphabet, gappy True) = Protein. Alphabet, gappy = True) -Protein_Alphabet, gappyTrue) gappy test_alnAlignment([seql, seq2, seq3, seq4, seq5]) print("My consensus:", test_aln.getConsensus ()) Submit (a): Use your getConsensus' to provide the consensus sequence for the five sequences above Above, as you nave seen in eaner practicals, s an image an alignment OT sequences. | ne alignment nignlignts tnose columns that are conserved. nis tells us which parts of the sequence are under the greatest evolutionary pressure. When a column is conserved, it is particularly interesting to know which symbol (amino acid or nucleic acid) is the most common, .e. the consensus. Here is a method for determining the consensus symbol for any column for an Alignment object (this method has already been implemented in the Alignment class). Please study it carefully; note that it does not care what the alphabet is so it counts the gap character ".just as any other character Note: self is a reference to the class instance 19 es. T es ite)e In [ 1: def getConsensusForColumn(self, colidx): for seq in self.seqs: mysym - seq[colidx] try: #Attempt to execute the code in the below block symcnt [mysym] 1 except: #If the above block of code throws any kind of error, # e.g. a dictionary key error, execute the below block of code symcnt [mysym]1 consensus None maxcnt0 for mysym in symcnt: if symcnt[mysym] maxcnt: maxcntsymcnt [mysym] consensus - mysym return consensus Hint: if you do not understand what the try and except code is doing, you can read more about it here Exercise 3 Write another method for the Alignment class called getConsensus. It should construct and return a neW Sequence from the consensus characters for each column in an alignment. You are recommended to repeatedly call getConsensusForColumn in your implementation. Note that the returned sequence should be composed from the same alphabet as the alignment. To test your code, use the following sequences and check your consensus sequence using Coder Quiz: In [ ] : = Sequence(name- "Seq1", sequence- "AGFDTVTTA?SWSL-YLVTNPNVOKKIO", alphabet = Sequence(name- "Seq2", sequence = "AGFDTITTAISWSL-YLVMNPR1QKK10", alphabet Sequence(name = "Seq3", sequence = "AGFDTVT-A-SWS LMYLVMNPRVORK10", alphabet = Sequence(name = "Seq4", sequence- "AGFDTVT-A-SWSLMYLVTNPRVORK10", alphabet Sequence (name "Seq5", sequence"AGFDTVT-AISWSLMYLVTNPGVQRKIQ", alphabet Protein-Alphabet, seq1 seq2 seq3 seq4 seq5 - True) Protein-Alphabet, gappy - True) Protein. Alphabet, gappy True) = Protein. Alphabet, gappy = True) -Protein_Alphabet, gappyTrue) gappy test_alnAlignment([seql, seq2, seq3, seq4, seq5]) print("My consensus:", test_aln.getConsensus ()) Submit (a): Use your getConsensus' to provide the consensus sequence for the five sequences above







