Question: def get_first_mutation_index(dna_string, generated_rna_string): dna_string: str generated_rna_string: str Returns int. Sometimes, errors occur during transcription that result in a different mRNA sequence from what is expected.
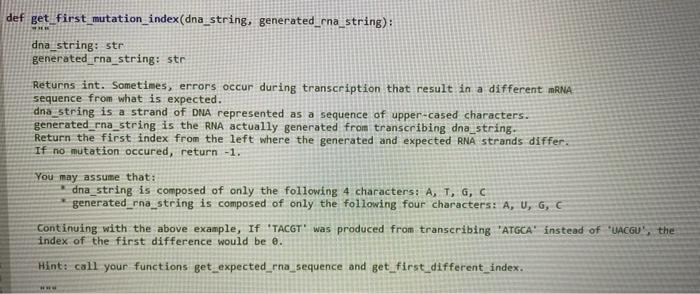
def get_first_mutation_index(dna_string, generated_rna_string): dna_string: str generated_rna_string: str Returns int. Sometimes, errors occur during transcription that result in a different mRNA sequence from what is expected. dna_string is a strand of DNA represented as a sequence of upper-cased characters. generated rna_string is the RNA actually generated from transcribing dna_string. Return the first index from the left where the generated and expected RNA strands differ If no mutation occured, return -1. You may assume that: dna_string is composed of only the following 4 characters: A, T, G, C generated_rna_string is composed of only the following four characters: A, U, G, C Continuing with the above example, If 'TACGT' was produced from transcribing "ATGCA' instead of 'UACGUS the index of the first difference would be e. Hint: call your functions get_expected_ena_sequence and get_first_different_index
Step by Step Solution
There are 3 Steps involved in it

Get step-by-step solutions from verified subject matter experts


