I am working on Rstudio, whenever I put a code is giving me an error, what might be the reason. > source(http://bioconductor.org/biocLite.R) > biocLite(SRAdb) >
I am working on Rstudio, whenever I put a code is giving me an error, what might be the reason. 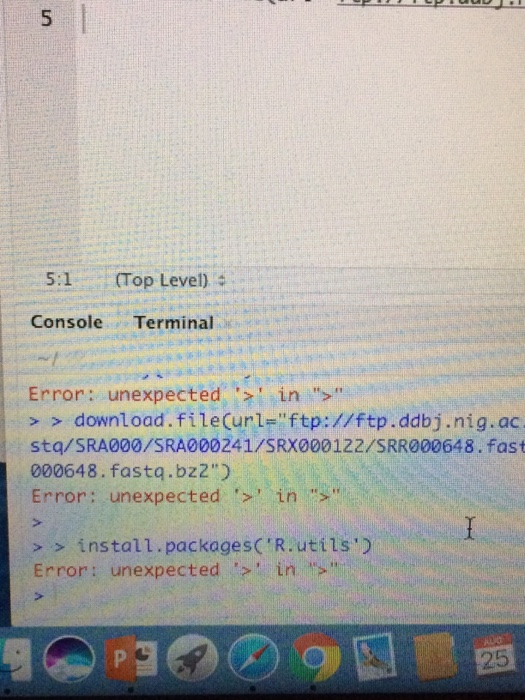
> source("http://bioconductor.org/biocLite.R") > biocLite("SRAdb") > library(SRAdb) > sqlFile sraCon install.packages("R.utils") > library(R.utils) > download.file(url="ftp://ftp.ddbj.nig.ac.jp/ddbj_database/dra/fastq/SRR926846/SRR926845.fastq.bz2") > bunzip2(list.files(pattern = ".fastq.bz2$")) > source("http://bioconductor.org/biocLite.R") > biocLite("ShortRead") > library(ShortRead)
these are my codes, I want to run them in R studio, put I could not. these codes are only to pring accession number from the SRA database, and the configure it to certain file type to start the gene expression analysis.
Also, I deleted files from my desktop through R, how can I retraive them.
5:1 Top Level) Console Terminal Error: unexpected in ">" > > download.fileCurl="ftp://ftp.ddbj.nig.ac stq/SRA000/SRA000241/SRX000122/SRR000648. fast 000648. fastq.bz2 Error: unexpectedin ">" > > install.packages('R.utils') Error: unexpected in >" 25Step by Step Solution
There are 3 Steps involved in it
Step: 1

See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


