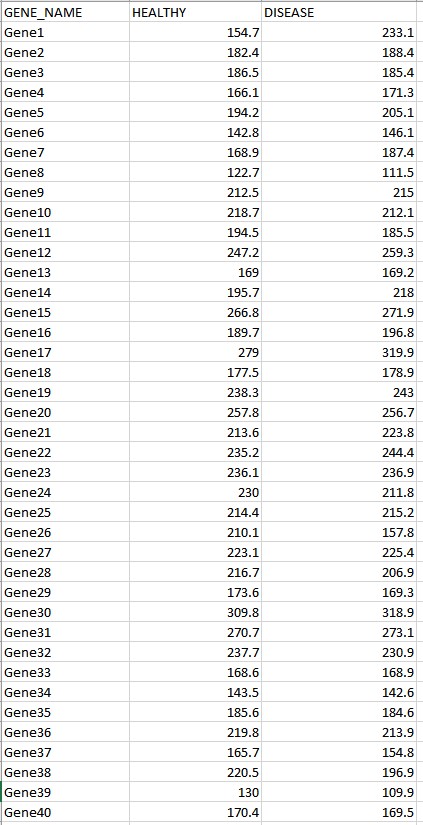
Introduction In this project we'll explore some of the techniques that could be used to explore gene ex\" pression activity. As a motivating example, we'll imagine that we are comparing the gene activity in a healthy human lung tissue sample to the gene activity in a lung cancer tumor sample. We will attempt to identify genes that seem to be showing a change in activity in tumor tissue samples relative to healthy control samples. A quick primer on gene expression: while we won't dive too deeply into the underlying biology, the basic idea is that genes (DNA sequences) in the genome are transcribed into messenger RNA sequences, which are then used as blueprints to manufacture proteins (the biological molecules which carry out most cellular functions). When we reference \"gene ach tivity\" in this study, we are broadly addressing this process of taking instructions from the genome and using them to produce proteins for the cell. Each gene will have a baseline, quantitative activity level in healthy (lung) tissue the genes may be inactive / off, or active to some extent following a spectrum of values corresponding to how much protein product is ultimately produced. The numerical value associated with \"activity\" broadly captures the number of messenger RNAs and,! or proteins produced. This activity level often changes dramatically in cancerous tissues, and the focus of this study will be to explore some tech niques that are used to identify and study the genes that are behaving dierently in tumors based on their observed, numerical activity levels- Note: Genes work in complicated ways, and you shouldn't read too much into the meaning of a positive difference in activity versus a negative difference in activity. A drop in activity could result in less production of some compound, or in less production of something inhibiting production of that compound.[ The study will proceed in two phases: First, we will compare the gene activity levels for 40 handpicked genes across two samples (one healthy, one cancerous). These genes are suspected to be broadly involved in tumorigenesis and the onset of cancer, but we'll seek to conrm whether these genes are behaving differently in this strain of cancer specically. To do this, we will use a scatterplot and regression analysis to explore the overall trend in gene activity behaviour (and to provide a preliminary glimpse into the genes that may be showing dilferences in activity level). In the second phase of the study, we'll collect multiple replicate measurements from healthy and disease tissues, allowing us to conduct hypothesis tests to conrm the (statistical) signicance of any changes in activity level. We will rst do this focusing on a subset of the 40 candidates (those that seemed most suspicious from the scatterplot / regression analysis), and then using the complete set of 40 candidates. Because we will be conducting several hypothesis tests in the context of a single study, this will give us a chance to test out corrections for the multiple testing problem. Scatterplot and Regression Analysis (3 gures expected) For this stage of the study we will be comparing the gene activity levels in one healthy tissue sample to the gene activity levels in one diseased (lung cancer) tissue sample. Each sample will provide one activity reading for each of the 40 genes included in our handpicked bundle of likely candidates for analysis. These expression levels are provided in the spreadsheet \"Regression Dataxls\". This spreadsheet has one row for each of the 40 included genes and two columns: one for the healthy tissue sample (column A] and one for the disease tissue sample [column B}. Question 1 a): (2 marks) Using lGeogebra (or a statistical software package of your choice), produce a sidebyside pair of boxplots, one for each of the two tissue samples being com pared. These boxplots will show the overall distribution of gene activity levels within each tissue. Please attach this boxplot as the rst gure of your report. (Geogebra Instructions: using the spreadsheet interface, read in the two columns of data, highlight all the data, and then select \"Multiple Variable Analysis\" using the set of buttons in the top left. By default, the rst visualization on the left should be a pair of stacked boxplots.) Question 1 b): (2 marks) The overall appearance of the two boxplots will be very similar. 1|Elfhat does this suggest about the two tissue samples? Comment on what it means for the boxplots to be similar, and discuss whether you believe this makes it impossible for any individual genes to be showing differences in activity levels between the two samples. [It might be helpful to review your answer to this problem later after inspecting a scatterplot.) Question ] c]: (3 marks} Using Geogebra [or a statistical software package of your choice}, produce a scatterplot comparing the values from healthy tissue (on the horizontal axis} to the values from diseased tissue {on the vertical axis). Include a linear regression line and provide the equation for the regression model. Please attach this scatterplot+regression line to your report as second gure of your report. [Geogebra Instructions: using the spread sheet interface, read in the two columns of data, highlight all the data, and then select \"Two Variable Regression Analysis\" using the set of buttons in the top left. By default, the rst visualization on the left should be a Scatterplot. At the bottom of this panel, you'll see a dropdown box that allows you to select a type of regression model. Select \"linear\GENE NAME HEALTHY DISEASE Gene1 154.7 233.1 Gene2 182.4 188.4 Gene3 186.5 185.4 Gene4 166.1 171.3 Gene5 194.2 205.1 Gene6 142.8 146.1 Gene7 168.9 187.4 Genes 122.7 111.5 Gene9 212.5 215 Gene10 218.7 212.1 Genel1 194.5 185.5 Gene12 247.2 259.3 Gene13 169 169.2 Gene14 195.7 218 Gene15 266.8 271.9 Gene16 189.7 196.8 Gene17 279 319.9 Gene18 177.5 178.9 Gene19 238.3 243 Gene20 257.8 256.7 Gene21 213.6 223.8 Gene22 235.2 244.4 Gene23 236.1 236.9 Gene24 230 211.8 Gene25 214.4 215.2 Gene26 210.1 157.8 Gene27 223.1 225.4 Gene28 216.7 206.9 Gene29 173.6 169.3 Gene30 309.8 318.9 Gene31 270.7 273.1 Gene32 237.7 230.9 Gene33 168.6 168.9 Gene34 143.5 142.6 Gene35 185.6 184.6 Gene36 219.8 213.9 Gene37 165.7 154.8 Gene38 220.5 196.9 Gene39 130 109.9 Gene40 170.4 169.5