Question
starter code #include #define TISSUE_SIZE (10) typedef enum tissueType { _ = '_', // Uninfected cell I = 'I', // Infected cell S = 'S',
starter code
#include
#define TISSUE_SIZE (10)
typedef enum tissueType { _ = '_', // Uninfected cell I = 'I', // Infected cell S = 'S', // Good bacterium: Sakinacoccus C = 'C', // Good bacterium: Charlestridium tetani } TissueType;
typedef struct cell { TissueType type; // Indicates if good bacterium, infected cell or uninfected cell int health; // For good bacteria: the multiple the range is extended by, for infected cells, // number of lives } Cell;
typedef Cell Tissue[TISSUE_SIZE][TISSUE_SIZE]; // A nice name for a 2D Cell array -> Tissue
/** * CRUNCHY: Use this helper helper function to implement Charles AND Sakina * HINT: I would implement numberOfCharlesInRange like this: * Pattern charles_patterns[4] = {{2, 0}, {0, 2}, {-2, 0}, {0, -2}}; * return numberOfBacteriaInRange(tissue_sample, x, y, charles_patterns, 4, C); */ typedef int Pattern[2]; int numberOfBacteriaInRange(Tissue tissue, int x, int y, Pattern pattern[], int num_patterns, TissueType type { for /** CRUNCHIER: do this without looping over all the cells (again) (attacks are symmetric!) */ int counter = 0;
return counter; }
/** * Return the number of Charlestridium tetani within tissue_sample that can attack position x, y. * * Legal attacks are limited to the pattern (where X is a legal attack and C is the bacterium): * _ _ X _ _ * _ _ _ _ _ * X _ C _ X * _ _ _ _ _ * _ _ X _ _ * where each Charlestridium tetani can attack in any one direction in this pattern for a certain * number of attacks. For example, if one had 2 health, its pattern would be limited to: * _ _ _ _ X _ _ _ _ * _ _ _ _ _ _ _ _ _ * _ _ _ _ X _ _ _ _ * _ _ _ _ _ _ _ _ _ * X _ X _ C _ X _ X * _ _ _ _ _ _ _ _ _ * _ _ _ _ X _ _ _ _ * _ _ _ _ _ _ _ _ _ * _ _ _ _ X _ _ _ _ * * INPUT: * tissue_sample: * - an array of arrays of Cells with dimensions TISSUE_SIZE * TISSUE_SIZE * - gives positions of each bacterium and each infected cell * - each cell can only contain one of the following types: * - `I`: Infected Cell * - `L`: A Charlestridium tetani bacterium * - `S`: A Sakinacoccus bacterium * - `_`: An uninfected cell (bacteria will ignore these) * x: the row coordinate (starting at 0) of the cell (within tissue_sample) to be checked. * y: the column coordinate (starting at 0) of the cell (within tissue_sample) to be checked. * * RETURN: the number of Charlestridium tetani that can make a legal attack to position x, y */ int numberOfCharlesInRange(Tissue tissue_sample, int x, int y) { /** TODO: implement this function, satisfying the description above. */
return 0; // replace this with your implementation }
/** * Return the number of Sakinacoccus within tissue_sample that can attack position x, y. * * Legal attacks are limited to the pattern (where X is a legal attack and S is the bacterium): * _ _ _ _ _ * _ X _ X _ * _ _ S _ _ * _ X _ X _ * _ _ _ _ _ * where each Sakinacoccus can attack in any one direction in this pattern for a certain * number of attacks. For example, if one had 2 health, its pattern would be limited to: * X _ _ _ X * _ X _ X _ * _ _ S _ _ * _ X _ X _ * X _ _ _ X * * INPUT: * tissue_sample: * - an array of arrays of Cells with dimensions TISSUE_SIZE * TISSUE_SIZE * - gives positions of each bacterium and each infected cell * - each cell can only contain one of the following types: * - `I`: Infected Cell * - `L`: A Charlestridium tetani bacterium * - `S`: A Sakinacoccus bacterium * - `_`: An uninfected cell (bacteria will ignore these) * x: the row coordinate (starting at 0) of the cell (within tissue_sample) to be checked. * y: the column coordinate (starting at 0) of the cell (within tissue_sample) to be checked. * * RETURN: the number of Sakinacoccus that can make a legal attack to position x, y */ int numberOfSakinaInRange(Tissue tissue_sample, int x, int y) { /** TODO: implement this function, satisfying the description above. */
return 0; // replace this with your implementation }
/** * Computes the percentage of infected cells in the given tissue sample which can be killed * by Sakinacoccus or Charlestridium tetani bacteria in the sample and updates the tissue sample to * the state after the attacks. * * INPUT: * tissue_sample: * - an array of arrays of Cells with dimensions TISSUE_SIZE * TISSUE_SIZE * - gives positions of each bacterium and each infected cell * - each cell can only contain one of the following types: * - `I`: Infected Cell * - `L`: A Charlestridium tetani bacterium * - `S`: A Sakinacoccus bacterium * - `_`: An uninfected cell (bacteria will ignore these) * - REQ: there is at least one infected cell * * OUTPUT: * tissue_sample: - mutated such that: * - All `I` cells have their health reduced by the number of good bacteria that can attack. * - Any `I` cell whose health drops to Think about how this 2D array is stored in memory... */
return 0; }
/** * Prints the tissue sample in a human readable format. * * INPUT: * tissue_sample: * - an array of arrays of Cells with dimensions TISSUE_SIZE * TISSUE_SIZE * - gives positions of each bacterium and each infected cell * - each cell can only contain one of the following types: * - `I`: Infected Cell * - `L`: A Charlestridium tetani bacterium * - `S`: A Sakinacoccus bacterium * - `_`: An uninfected cell (bacteria will ignore these) * * NOTE: The displayed indices do not match the implementation. * That is, they start at 1 (when we know array indices start at 0). * This is more obvious for the column indices, which are A - J. */ void printTissueSample(Tissue tissue_sample) { printf(" A B C D E F G H I J "); for (int x = 0; x
// We NEED this compiler directive to test your code with our own main(). Don't break it. #ifndef __TESTING // this compiler directive int main() { // Please note that these are JUST EXAMPLES! You will be tested on a wide range of data.
printf("====Jackson's Sample==== "); Tissue jackson_sample = { {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {C, 1}, {_, 0}, {I, 1}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 3}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 2}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {S, 2}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}}; printTissueSample(jackson_sample); printf("Calculated: Killed %06.2f%% of bacteria ", testBacteria(jackson_sample)); printf(" Expected: Killed 066.67%% of bacteria "); printTissueSample(jackson_sample);
// Feel free to add your own examples/tests here printf("====Silviu's Sample==== "); Tissue silviu_sample = { {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}, {_, 0}, {_, 0}, {I, 1}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}}, {{_, 0}, {_, 0}, {_, 0}, {I, 1}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}, {S, 2}}, {{_, 0}, {_, 0}, {_, 0}, {I, 1}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {C, 3}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {C, 2}, {_, 0}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}, {I, 1}, {I, 1}}, {{_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {_, 0}, {I, 1}, {_, 0}}}; printTissueSample(silviu_sample); printf("Calculated: Killed %06.2f%% of bacteria ", testBacteria(silviu_sample)); printf(" Expected: Killed 036.36%% of bacteria "); printTissueSample(silviu_sample); }
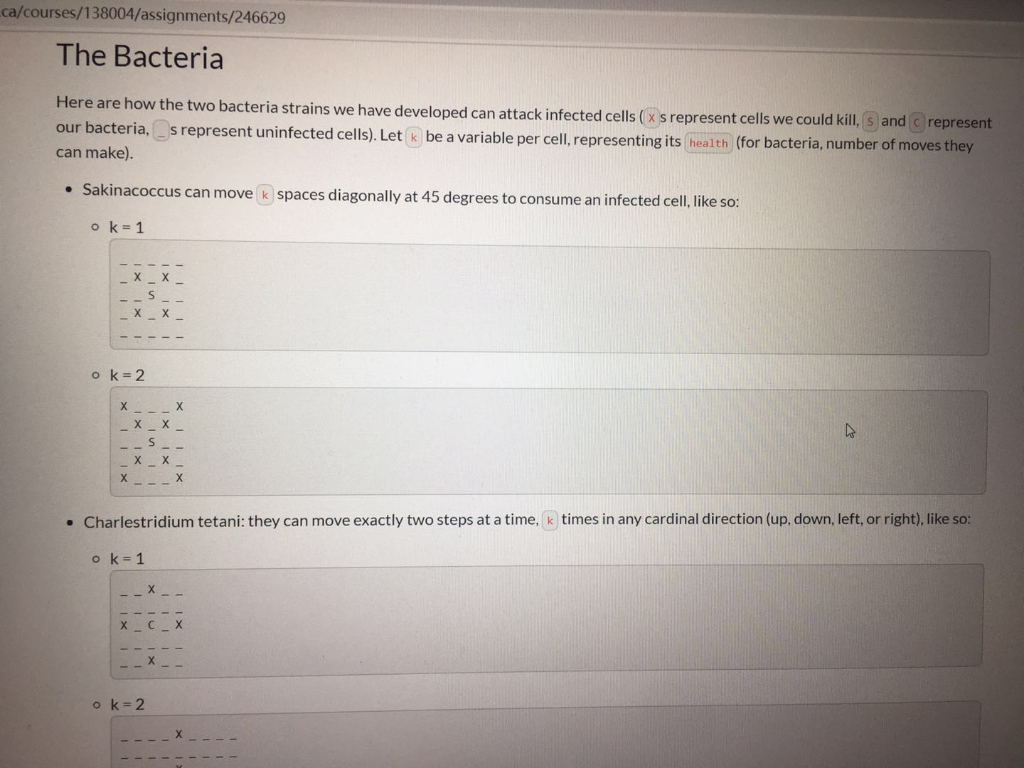
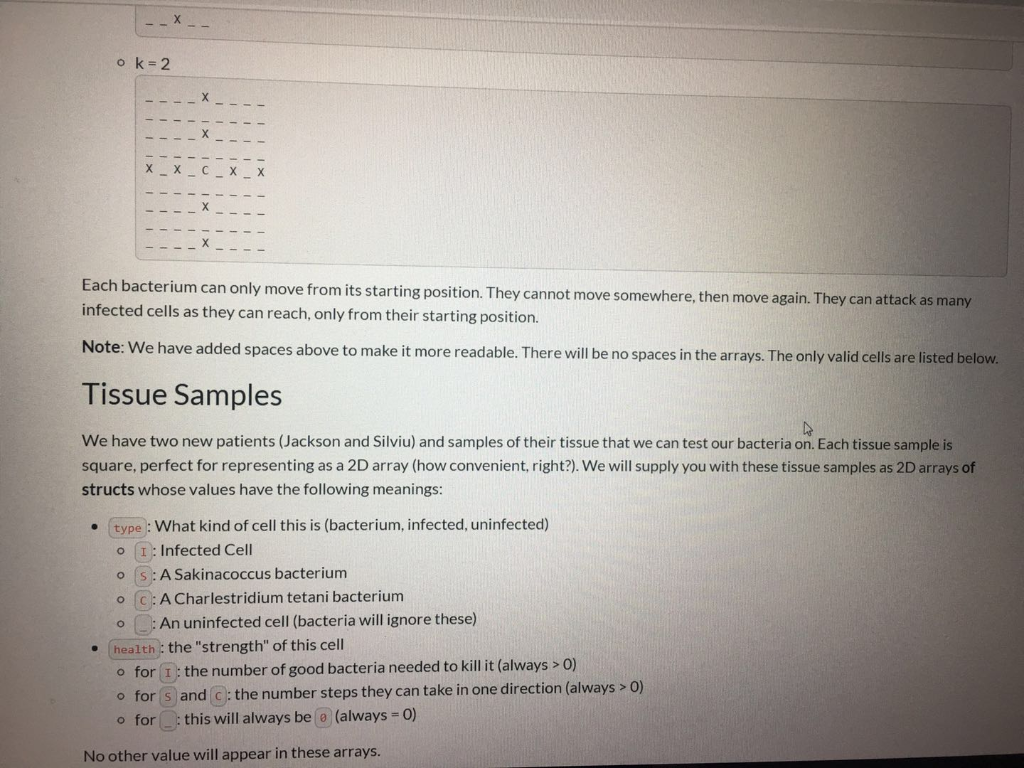
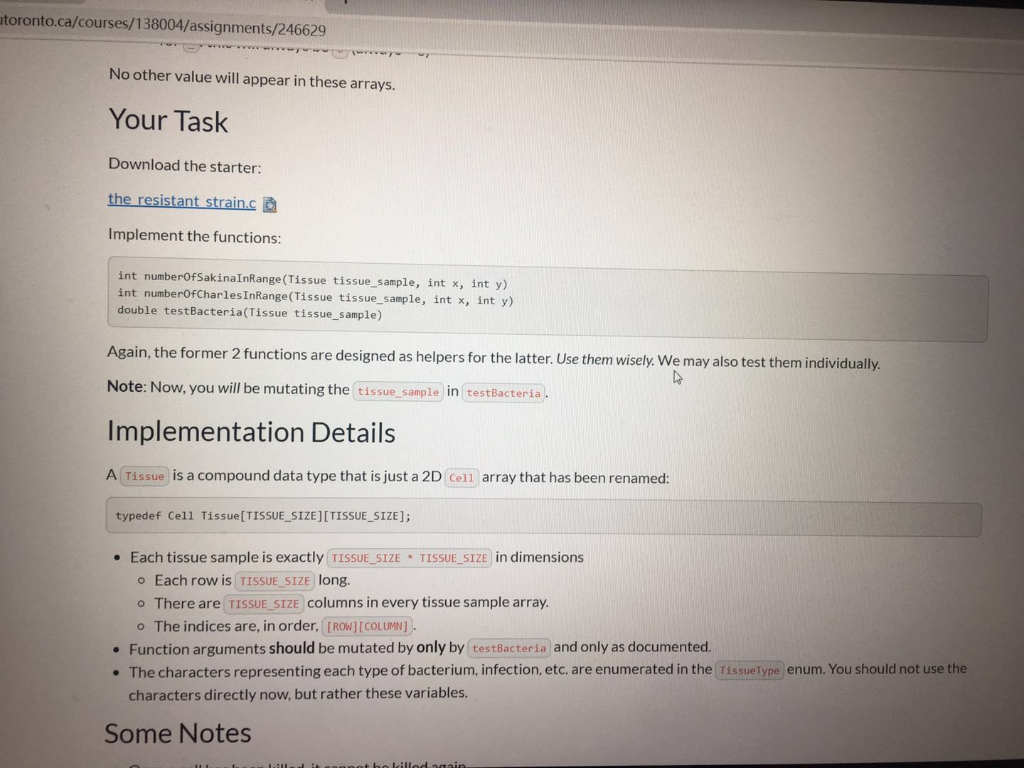
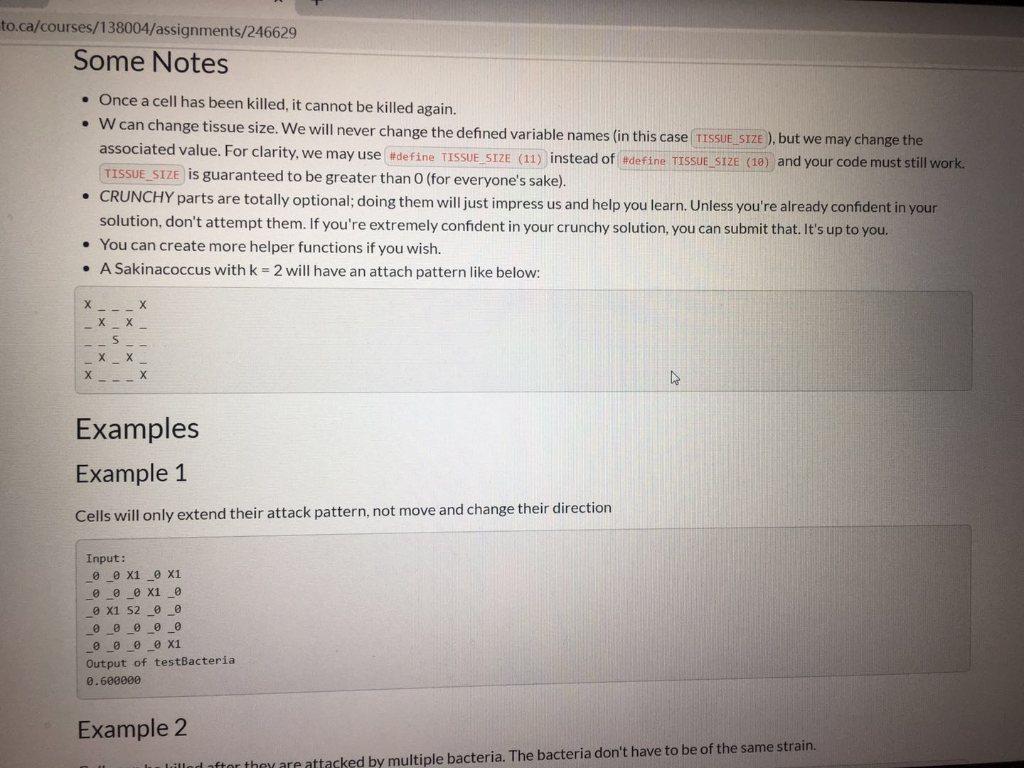
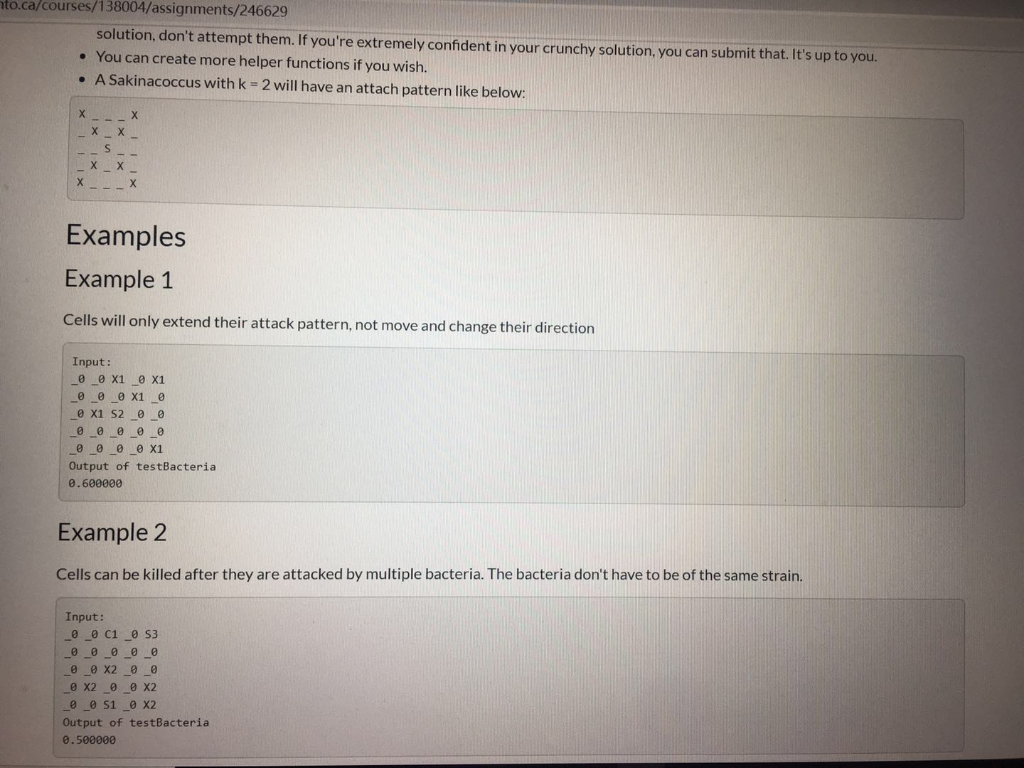
ca/courses/138004/assignments/246629 The Bacteria Here are how the two bacteria strains we have developed can attack infected cells ( x s represent cells we could kill, s and c represent our bacteria, _ s represent uninfected cells). Let k be a variable per cell, representing its health (for bacteria, number of moves they can make). Sakinacoccus can move k spaces diagonally at 45 degrees to consume an infected cell, like so: o k=1 1 X TXI XI 1 X 1 ok=2 1 X Tin X II 1X | XI XI x | x 1 X ! Charlestridium tetani: they can move exactly two steps at a time, k times in any cardinal direction (up, down, left, or right), like so: ok=1 II X 11 1 1 1 1 X 11 1 1 ok=2 --X - ok=2 III IX III X 1 I X 1 X TIL TIL 10 TILL II III IX II IIIIIIIII III IX III X 1 X 1 Each bacterium can only move from its starting position. They cannot move somewhere, then move again. They can attack as many infected cells as they can reach, only from their starting position. Note: We have added spaces above to make it more readable. There will be no spaces in the arrays. The only valid cells are listed below. Tissue Samples We have two new patients (Jackson and Silviu) and samples of their tissue that we can test our bacteria on. Each tissue sample is square, perfect for representing as a 2D array (how convenient, right?). We will supply you with these tissue samples as 2D arrays of structs whose values have the following meanings: type : What kind of cell this is (bacterium, infected, uninfected) O I:Infected Cell OS: A Sakinacoccus bacterium OC: A Charlestridium tetani bacterium O : An uninfected cell (bacteria will ignore these) health : the "strength" of this cell o for I : the number of good bacteria needed to kill it (always > 0) o for s and c: the number steps they can take in one direction (always > 0) o for this will always be (always=0) No other value will appear in these arrays. utoronto.ca/courses/138004/assignments/246629 No other value will appear in these arrays. Your Task Download the starter: the resistant strain.ca Implement the functions: int numberOfSakinaInRange(Tissue tissue_sample, int x, int y) int numberOfCharles InRange(Tissue tissue_sample, int x, int y) double testBacteria(Tissue tissue_sample) Again, the former 2 functions are designed as helpers for the latter. Use them wisely. We may also test them individually. Note: Now, you will be mutating the tissue_sample in testBacteria. Implementation Details A Tissue is a compound data type that is just a 2D Cell array that has been renamed: typedef Cell Tissue[TISSUE_SIZE][TISSUE_SIZE]; Each tissue sample is exactly TISSUE_SIZE TISSUE_SIZE in dimensions o Each row is TISSUE_SIZE long. There are TISSUE_SIZE columns in every tissue sample array. o The indices are, in order, [ROW] [COLUMN]. Function arguments should be mutated by only by testBacteria and only as documented. The characters representing each type of bacterium, infection, etc. are enumerated in the Tissue Type enum. You should not use the characters directly now, but rather these variables. Some Notes Lill e lilled to.ca/courses/138004/assignments/246629 Some Notes Once a cell has been killed, it cannot be killed again. . W can change tissue size. We will never change the defined variable names in this case TISSUE_SIZE ), but we may change the associated value. For clarity, we may use #define TISSUE_SIZE (11) instead of #define TISSUE SIZE (10) and your code must still work. TISSUE_SIZE is guaranteed to be greater than 0 (for everyone's sake). CRUNCHY parts are totally optional; doing them will just impress us and help you learn. Unless you're already confident in your solution, don't attempt them. If you're extremely confident in your crunchy solution, you can submit that. It's up to you. You can create more helper functions if you wish. A Sakinacoccus with k = 2 will have an attach pattern like below: I | 3 1X X II IX X II IX | 4 in 11 1X | 1 Examples Example 1 Cells will only extend their attack pattern, not move and change their direction Input: _ _ x1 _ x1 _ _ _e x1 _ @ x1 52 _ _ _@0_0_0_0 O O O X1 Output of testBacteria 0.600000 Example 2 In lilled after they are attacked by multiple bacteria. The bacteria don't have to be of the same strain ato.ca/courses/138004/assignments/246629 solution, don't attempt them. If you're extremely confident in your crunchy solution, you can submit that. It's up to you. You can create more helper functions if you wish. A Sakinacoccus with k = 2 will have an attach pattern like below: in 11 X II IX Examples Example 1 Cells will only extend their attack pattern, not move and change their direction Input: _ _ x1 _0 x1 000 x1 o x1 52 _ _ 0 0 0 0 0 @ @ @ @ X1 Output of testBacteria 0.600000 Example 2 Cells can be killed after they are attacked by multiple bacteria. The bacteria don't have to be of the same strain. Input: __O C1 _053 ooooo 0 0 x2 oo @ X2_0_X2 @ _@ 51 _ X2 Output of testBacteria 0.500000 ca/courses/138004/assignments/246629 The Bacteria Here are how the two bacteria strains we have developed can attack infected cells ( x s represent cells we could kill, s and c represent our bacteria, _ s represent uninfected cells). Let k be a variable per cell, representing its health (for bacteria, number of moves they can make). Sakinacoccus can move k spaces diagonally at 45 degrees to consume an infected cell, like so: o k=1 1 X TXI XI 1 X 1 ok=2 1 X Tin X II 1X | XI XI x | x 1 X ! Charlestridium tetani: they can move exactly two steps at a time, k times in any cardinal direction (up, down, left, or right), like so: ok=1 II X 11 1 1 1 1 X 11 1 1 ok=2 --X - ok=2 III IX III X 1 I X 1 X TIL TIL 10 TILL II III IX II IIIIIIIII III IX III X 1 X 1 Each bacterium can only move from its starting position. They cannot move somewhere, then move again. They can attack as many infected cells as they can reach, only from their starting position. Note: We have added spaces above to make it more readable. There will be no spaces in the arrays. The only valid cells are listed below. Tissue Samples We have two new patients (Jackson and Silviu) and samples of their tissue that we can test our bacteria on. Each tissue sample is square, perfect for representing as a 2D array (how convenient, right?). We will supply you with these tissue samples as 2D arrays of structs whose values have the following meanings: type : What kind of cell this is (bacterium, infected, uninfected) O I:Infected Cell OS: A Sakinacoccus bacterium OC: A Charlestridium tetani bacterium O : An uninfected cell (bacteria will ignore these) health : the "strength" of this cell o for I : the number of good bacteria needed to kill it (always > 0) o for s and c: the number steps they can take in one direction (always > 0) o for this will always be (always=0) No other value will appear in these arrays. utoronto.ca/courses/138004/assignments/246629 No other value will appear in these arrays. Your Task Download the starter: the resistant strain.ca Implement the functions: int numberOfSakinaInRange(Tissue tissue_sample, int x, int y) int numberOfCharles InRange(Tissue tissue_sample, int x, int y) double testBacteria(Tissue tissue_sample) Again, the former 2 functions are designed as helpers for the latter. Use them wisely. We may also test them individually. Note: Now, you will be mutating the tissue_sample in testBacteria. Implementation Details A Tissue is a compound data type that is just a 2D Cell array that has been renamed: typedef Cell Tissue[TISSUE_SIZE][TISSUE_SIZE]; Each tissue sample is exactly TISSUE_SIZE TISSUE_SIZE in dimensions o Each row is TISSUE_SIZE long. There are TISSUE_SIZE columns in every tissue sample array. o The indices are, in order, [ROW] [COLUMN]. Function arguments should be mutated by only by testBacteria and only as documented. The characters representing each type of bacterium, infection, etc. are enumerated in the Tissue Type enum. You should not use the characters directly now, but rather these variables. Some Notes Lill e lilled to.ca/courses/138004/assignments/246629 Some Notes Once a cell has been killed, it cannot be killed again. . W can change tissue size. We will never change the defined variable names in this case TISSUE_SIZE ), but we may change the associated value. For clarity, we may use #define TISSUE_SIZE (11) instead of #define TISSUE SIZE (10) and your code must still work. TISSUE_SIZE is guaranteed to be greater than 0 (for everyone's sake). CRUNCHY parts are totally optional; doing them will just impress us and help you learn. Unless you're already confident in your solution, don't attempt them. If you're extremely confident in your crunchy solution, you can submit that. It's up to you. You can create more helper functions if you wish. A Sakinacoccus with k = 2 will have an attach pattern like below: I | 3 1X X II IX X II IX | 4 in 11 1X | 1 Examples Example 1 Cells will only extend their attack pattern, not move and change their direction Input: _ _ x1 _ x1 _ _ _e x1 _ @ x1 52 _ _ _@0_0_0_0 O O O X1 Output of testBacteria 0.600000 Example 2 In lilled after they are attacked by multiple bacteria. The bacteria don't have to be of the same strain ato.ca/courses/138004/assignments/246629 solution, don't attempt them. If you're extremely confident in your crunchy solution, you can submit that. It's up to you. You can create more helper functions if you wish. A Sakinacoccus with k = 2 will have an attach pattern like below: in 11 X II IX Examples Example 1 Cells will only extend their attack pattern, not move and change their direction Input: _ _ x1 _0 x1 000 x1 o x1 52 _ _ 0 0 0 0 0 @ @ @ @ X1 Output of testBacteria 0.600000 Example 2 Cells can be killed after they are attacked by multiple bacteria. The bacteria don't have to be of the same strain. Input: __O C1 _053 ooooo 0 0 x2 oo @ X2_0_X2 @ _@ 51 _ X2 Output of testBacteria 0.500000Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


