Answered step by step
Verified Expert Solution
Question
1 Approved Answer
This assignment has a DNA_Strand.h. file that contains the declaration of a set of functions to manipulate static arrays representing DNA. There is another initial
This assignment has a DNA_Strand.h. file that contains the declaration of a set of functions to manipulate static arrays representing DNA. There is another initial test program: DNAtest.cpp. The assignment is to add code to the DNAtest.cpp file to fully test the DNA_Strand class to ensure it works as expected. The task is to only test the DNA_Strand class, not to implement the DNA_Strand class. All the functions that need to be tested are fully described in the DNA_Strand.h file.
DNA_Strand.h.
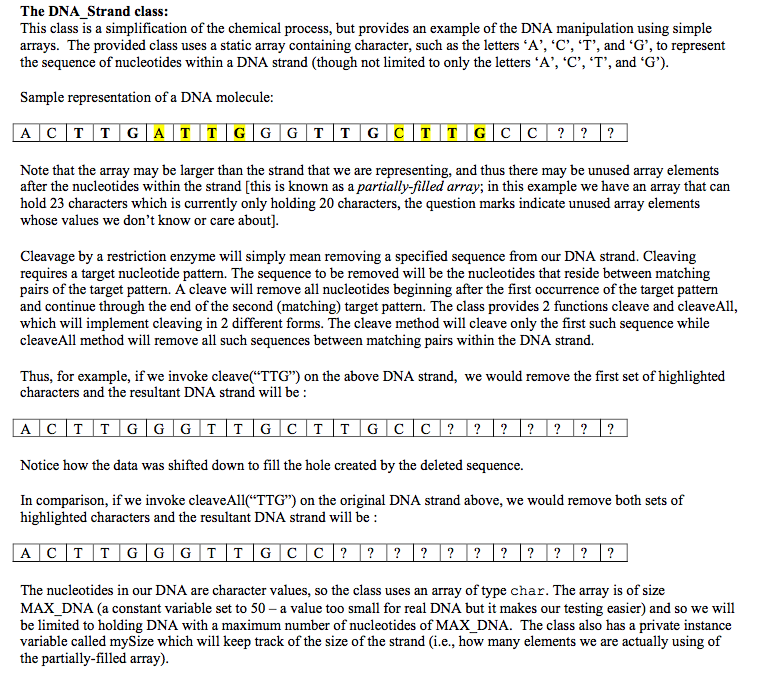
#ifndef DNA_STRAND_H #define DNA_STRAND_H #include//Size of static DNA Array const size_t MAX_DNA = 50; class DNA_Strand { private: // Returns true if index is within range of the current DNA_Strand // else returns false. bool inRange (size_t index) const; // Current size of the DNA_Strand; i.e., how many elements of the array are // currently being used (since this is a partially-filled array) size_t mySize; // DNA_Strand's storage buffer. May only be partially filled. char myDNA[MAX_DNA]; public: // Create an empty DNA_Strand. DNA_Strand (); // default ctor // Create an initialized DNA_Strand. // If the string ipStr is larger than MAX_DNA, then only the first MAX_DNA // characters of ipStr are used to initialize the DNA_Strand DNA_Strand (const std::string & ipStr); // ctor that takes a string //toString //Returns string equivalent of the DNA std::string toString() const; // Set an item in the DNA_Strand at location index. Throws // <:out_of_range> if index is out of range, i.e., larger than the // current size of the DNA_Strand. // Uses zero-based indexing. void set (char new_item, size_t index); // Get an item in the DNA_Strand at location index. Throws // <:out_of_range> if index is out of range, i.e., larger than the // current size of the DNA_Strand. // Uses zero-based indexing. char get (size_t index) const; // Returns the current size of the DNA. size_t size () const; // Compare this DNA_Strand with s for equality. Returns true if the // size()'s of the two DNA_Strands are equal and all the elements from 0 // .. size()-1 are equal, else false. bool isEqual(const DNA_Strand & s) const; // search // Look for target in current DNA strand and return index. // Return -1 if not found. int search(const std::string & target) const; // search with start position specified // Look for target in current DNA strand and return index. // Return -1 if not found. If pos is past end of strand, return -1. int search(size_t pos, const std::string & target) const; // cleave // Removes from current DNA strand the sequence between the end of the // first occurrence of passed target sequence (e.g. "TTG"), through the end // of the second occurence of the target sequence. // pre: Array e.g. ACTTGACCTTGA and target e.g. "TTG" // post: ACTTGA (ACCTTG removed) void cleave(const std::string & target); // cleave with start position specified // cleave starting from an index, returns the index after the cleaved DNA // or -1 if no cleaving was performed // pre: Array e.g. ACTTGACCTTGA and target e.g. "TTG", pos = 1 // post: ACTTGA (ACCTTG removed) and return value = 5 int cleave(size_t pos, const std::string & target); // cleaveAll // Removes from current DNA strand the sequence between pairs of target // sequence, i.e. from the end 1 through the end of 2, from the end of 3 // through the end of 4, etc, but NOT from the end of 2 through the end 3, // or from the end of 4 through the end of 5. // (Make sure that you understand the specification) // pre: Array e.g. ACTTGATTGGGTTGCTTGCC and target e.g. "TTG" // post: ACTTGGGTTGCC (ATTG and CTTG removed) void cleaveAll(const std::string & target); // countEnzyme // Counts the number of occurences of a single character target sequence // in the DNA strand. size_t countEnzyme(char target) const; }; #endif /* ifndef */
DNAtest.cpp.
#includeThe DNA Strand class: This class is a simplification of the chemical process, but provides an example of the DNA manipulation using simple arrays. The provided class uses a static array containing character, such as the letters A', C','T', and 'G", to represent the sequence of nucleotides within a DNA strand (though not limited to only the letters*A','C','T', and 'G') Sample representation of a DNA molecule Note that the array may be larger than the strand that we are representing, and thus there may be unused array elements after the nucleotides within the strand [this is known as a partially-filled array; in this example we have an array that can hold 23 characters which is currently only holding 20 characters, the question marks indicate unused array elements whose values we don't know or care about] Cleavage by a restriction enzyme will simply mean removing a specified sequence from our DNA strand. Cleaving requires a target nucleotide pattern. The sequence to be removed will be the nucleotides that reside between matching pairs of the target pattern. A cleave will remove all nucleotides beginning after the first occurrence of the target pattern and continue through the end of the second (matching) target pattern. The class provides 2 functions cleave and cleaveAl, which will implement cleaving in 2 different forms. The cleave method will cleave only the first such sequence while cleaveAll method will remove all such sequences between matching pairs within the DNA strand. Thus, for example, if we invoke cleave("TTG") on the above DNA strand, we would remove the first set of highlighted characters and the resultant DNA strand will be Notice how the data was shifted down to fill the hole created by the deleted sequence. In comparison, if we invoke cleaveAll("TTG") on the original DNA strand above, we would remove both sets of highlighted characters and the resultant DNA strand will be A CTT G G G T T G C C? The nucleotides in our DNA are character values, so the class uses an array of type char. The array is of size MAX_DNA (a constant variable set to 50 -a value too small for real DNA but it makes our testing easier) and so we will be limited to holding DNA with a maximum number of nucleotides of MAX_DNA. The class also has a private instance variable called mySize which will keep track of the size of the strand (i.e., how many elements we are actually using of the partially-filled array)#include #include "DNA_Strand.h" int main() { // testing functions of the DNA_Strand class DNA_Strand dna_1, dna_2; // create 2 default-constructed strands if (dna_1.size() != 0) { std::cout if (!dna_1.isEqual(dna_2)) { std::cout if (!dna_2.isEqual(dna_1)) { std::cout if (dna.toString() != ipStr) { std::cout try { std::cout catch (std::out_of_range& excpt) { std::cout catch (...) { std::cout return 0; }
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


