Answered step by step
Verified Expert Solution
Question
1 Approved Answer
use the sum of pair score instead of distance score and make the necessary adjustmints,please solve it in details and show all the steps: 4
use the sum of pair score instead of distance score and make the necessary adjustmints,please solve it in details and show all the steps:
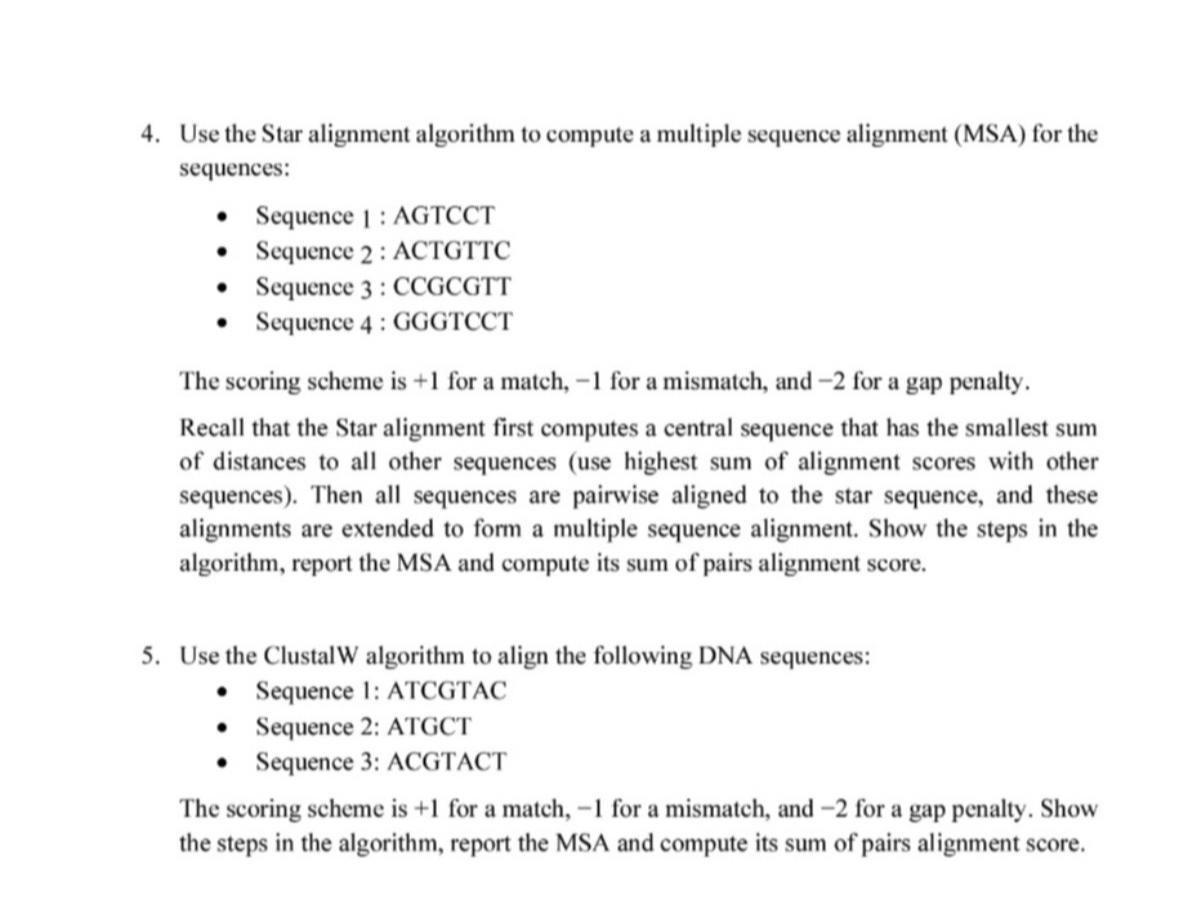
Use the Star alignment algorithm to compute a multiple sequence alignment MSA for the sequences:
Sequence : AGTCCT
Sequence : ACTGTTC
Sequence : CCGCGTT
Sequence : GGGTCCT
The scoring scheme is for a match, for a mismatch, and for a gap penalty.
Recall that the Star alignment first computes a central sequence that has the smallest sum of distances to all other sequences use highest sum of alignment scores with other sequences Then all sequences are pairwise aligned to the star sequence, and these alignments are extended to form a multiple sequence alignment. Show the steps in the algorithm, report the MSA and compute its sum of pairs alignment score.
use the ClustalW algorithm to align the following DNA sequences:
Sequence : ATCGTAC
Sequence : ATGCT
Sequence : ACGTACT
The scoring scheme is for a match, for a mismatch, and for a gap penalty. Show the steps in the algorithm, report the MSA and compute its sum of pairs alignment score.
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


