Answered step by step
Verified Expert Solution
Question
1 Approved Answer
why? DNA N-Gram Distribution (string scalar) In DNA analysis unique sequences of three bases need to be identified any point in the gene sequence. The
why?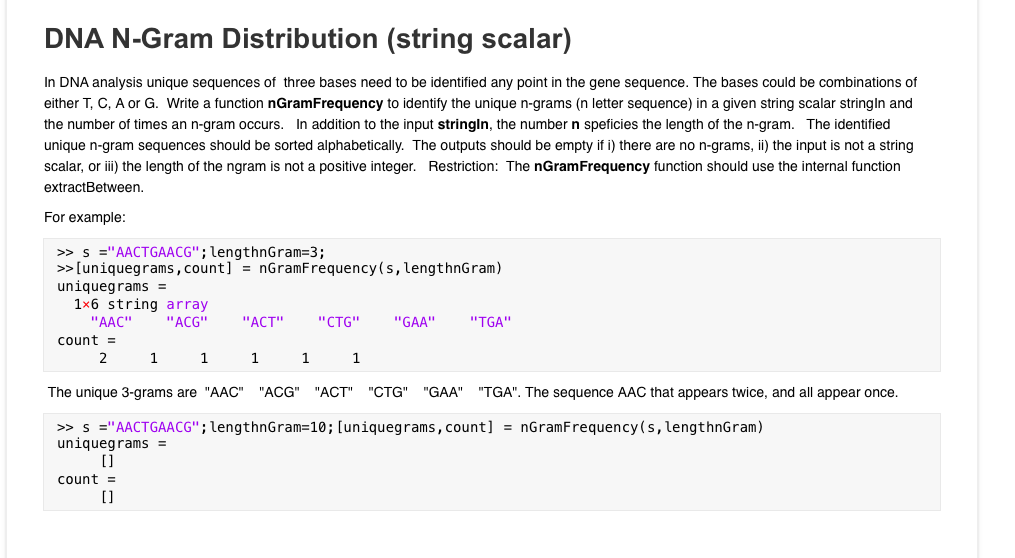
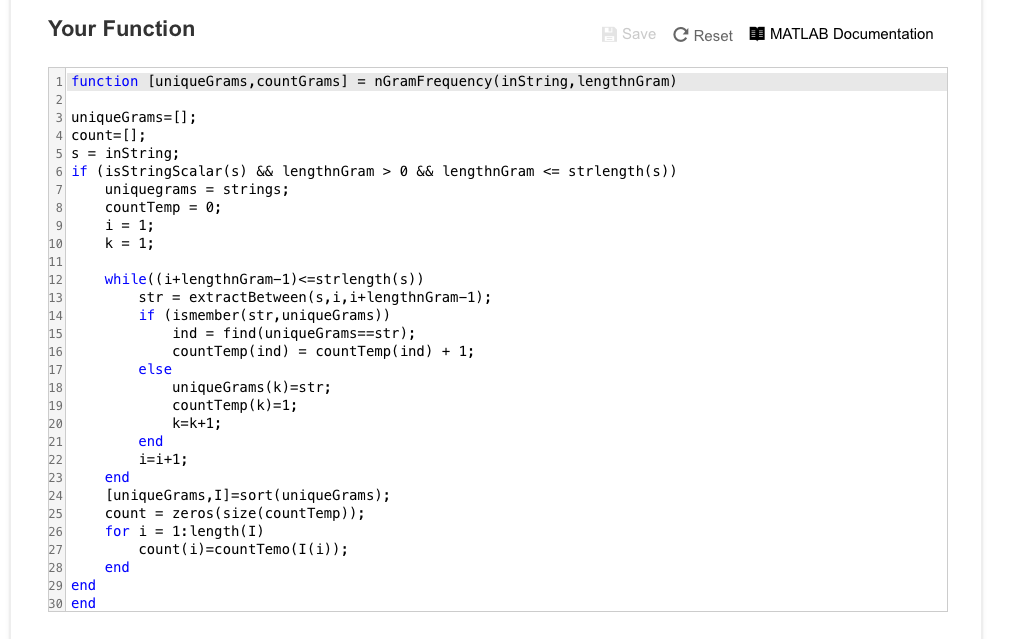
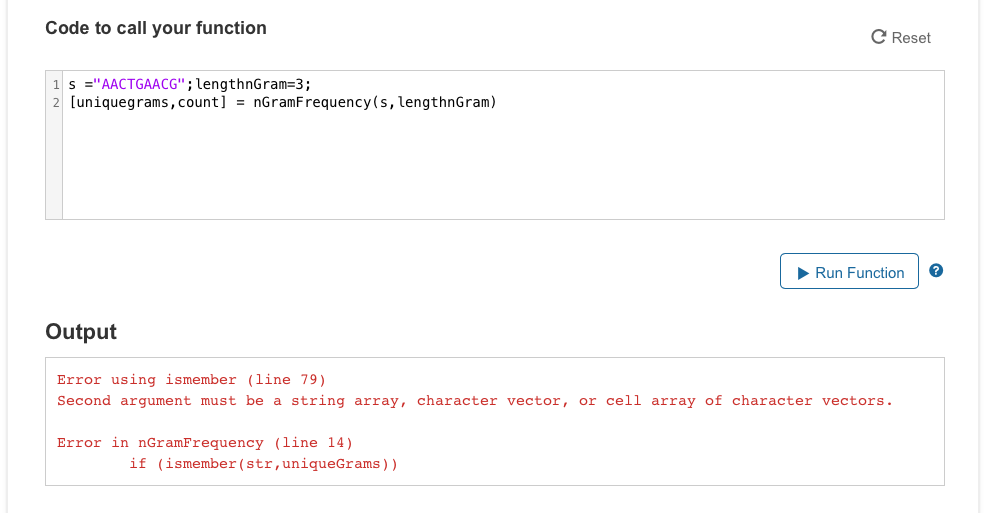
Step by Step Solution
There are 3 Steps involved in it
Step: 1

Get Instant Access to Expert-Tailored Solutions
See step-by-step solutions with expert insights and AI powered tools for academic success
Step: 2

Step: 3

Ace Your Homework with AI
Get the answers you need in no time with our AI-driven, step-by-step assistance
Get Started


